Interface between 40S exit channel protein uS7/Rps5 and eIF2α modulates start codon recognition in vivo
Figures
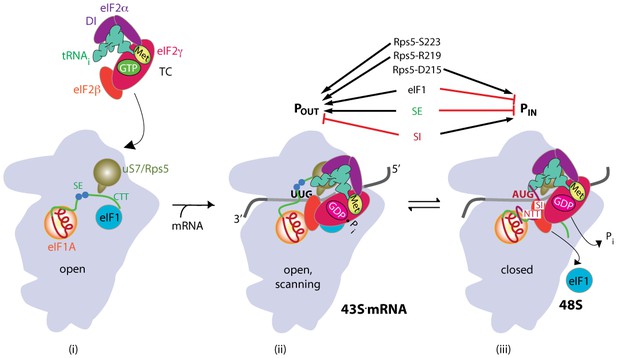
Model describing conformational rearrangements of the PIC during scanning and start codon recognition.
(i) eIF1 and the scanning enhancers (SEs) in the CTT of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly binds. uS7 is located in the mRNA exit channel of the 40S; (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAi bound in the POUT state and uS7 interacting with eIF2α-D1. eIF2 can hydrolyze GTP to GDP•Pi, but release of Pi is blocked. (iii) On AUG recognition, Met-tRNAi moves from the POUT to PIN state, clashing with eIF1 and the CTT of eIF1A, provoking displacement of the eIF1A CTT from the P site, dissociation of eIF1 from the 40S subunit, and Pi release from eIF2. The NTT of eIF1A, harboring scanning inhibitor (SI) elements, adopts a defined conformation and interacts with the codon:anticodon helix. The eIF2α-D1/uS7 interface is remodeled. (Above) Arrows summarize that eIF1 and the eIF1A SE elements promote POUT and impede transition to PIN state, whereas the scanning inhibitor (SI) element in the NTT of eIF1A stabilizes the PIN state. Results presented below indicate that uS7/Rps5 residue D215 promotes the closed conformation, whereas R219 and S223 enhance the open state (Adapted from Hinnebusch, 2014).
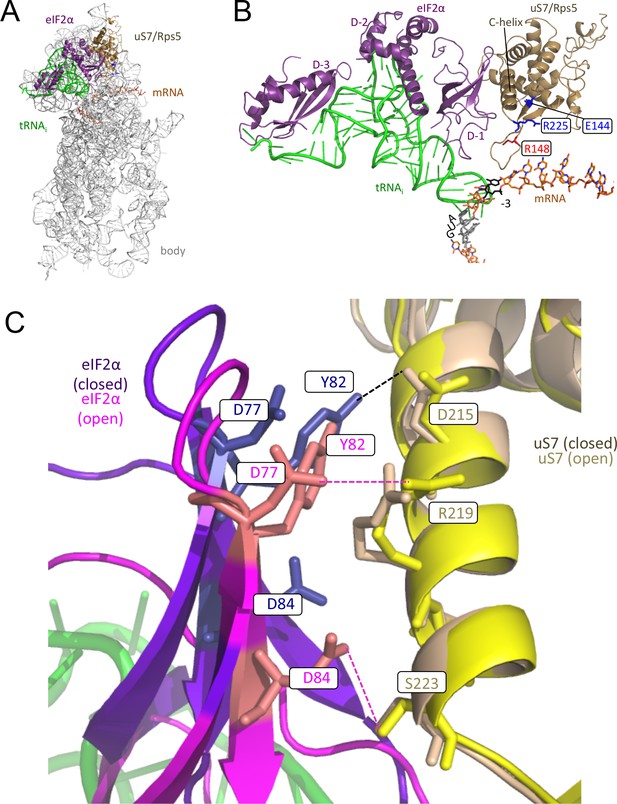
Alteration of the interface between eIF2α-D1 and C-terminal helix of uS7 in the open versus closed conformations of the py48S PIC.
(A, B) Depiction of the py48S PIC (PDB 3J81) showing uS7/Rps5 (gold), mRNA (orange), Met-tRNAi (green), eIF2α (purple). For clarity, other ribosomal proteins, eIF2β, eIF2γ, eIF1, eIF1A and putative eIF5 densities are not shown. uS7 residues previously implicated in promoting AUG recognition (Visweswaraiah et al., 2015) are shown in blue or red with stick side-chains. (C) Overlay of py48S-open (PDB 3JAQ) and py48S-closed (PDB 3JAP) revealing remodeling of the interface between eIF2α-D1 (purple or dark blue-closed complex; magenta or orange-open complex) and C-terminal helix of uS7 (beige-closed, yellow-open). Residues making contacts that appear to be favored in the open or closed state are shown with stick side-chains, using dotted lines to indicate the favored interactions.
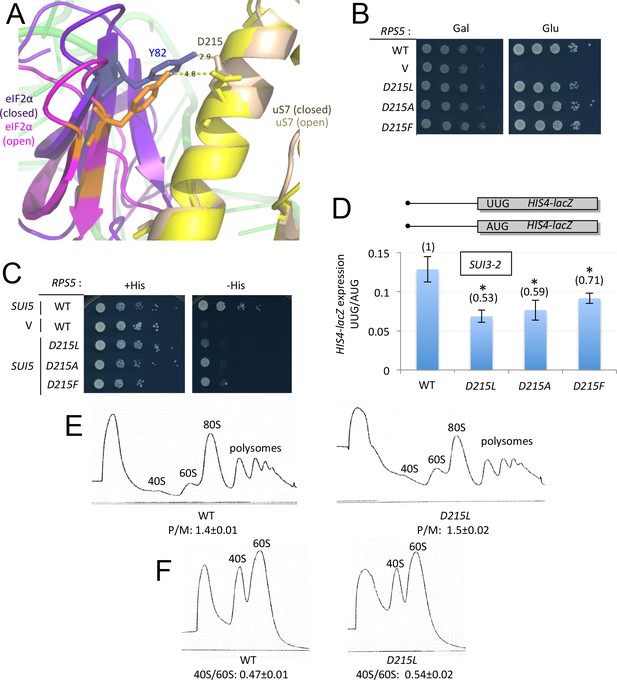
uS7-D215 substitutions increase discrimination against UUG start codons in vivo.
(A) Overlay of py48S-open and py48S-closed as in Figure 2C, showing that uS7-D215/eIF2α-Y82 interaction is favored in the closed complex (dark blue/beige sticks). (B) 10-fold serial dilutions of transformants of pGAL1-RPS5 his4–301 strain (JVY07) with the indicated plasmid-borne RPS5 alleles, or empty vector (V) were spotted on SCGal-Leu (Gal) or SC-Leu (Glu) and incubated at 30°C for 2–3 days. (C) 10-fold serial dilutions of JVY07 transformants with the indicated RPS5 alleles and SUI5 plasmid p4281, or empty vector (V) were spotted on SD+Ura+His (+His) or SD+Ura (−His) and incubated at 30°C for 3d and 5d, respectively. (D) JVY07 transformants with the indicated RPS5 alleles, SUI3–2 plasmid p4280, and HIS4-lacZ reporters with AUG or UUG start codons (plasmids p367 and p391, respectively) were cultured in SD+His at 30°C to an A600 of ~1 and β-galactosidase specific activities were measured in WCEs in units of nanomoles of o-nitrophenyl-β-D-galactopyranoside (ONPG) cleaved per min per mg of total protein. Ratios of mean expression of the UUG and AUG reporters calculated from four biological and two technical replicates are plotted with error bars (indicating S.E.M.s). *p<0.05 (E) WT and JVY76 (rps5-D215L) were cultured in SC-Leu at 30°C to A600 of ~1, and cycloheximide was added prior to harvesting. WCEs were separated by sucrose density gradient centrifugation and scanned at 254 nm to yield the tracings shown. Mean polysome/monosome ratios (and S.E.M.s) from three biological replicates are indicated. (F) Similar to (E) but cultures were not treated with cycloheximide and lysed in buffers without MgCl2 to allow separation of dissociated 40S and 60S ribosomal subunits. Mean 40S/60S ratios (and S.E.M.s) from three biological replicates are indicated.
-
Figure 3—source data 1
Effects of Rps5-D215 substitutions on HIS4-lacZ UUG:AUG expression ratios and polysome:monosome ratios.
- https://doi.org/10.7554/eLife.22572.005
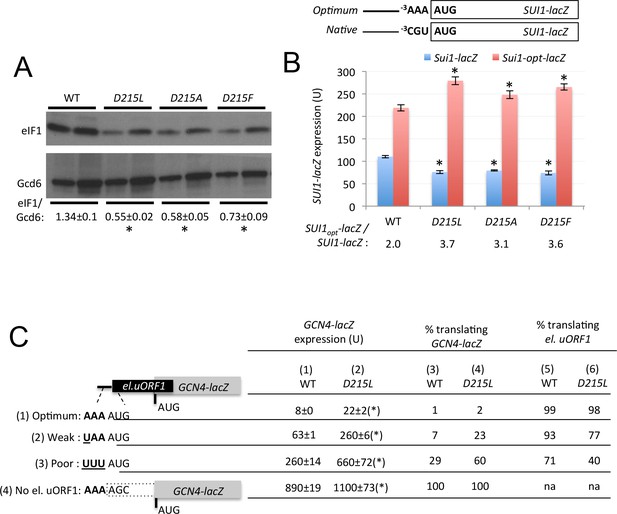
uS7 substitution D215L discriminates against AUG start codons in poor context.
(A) WCEs of strains from Figure 3B subjected to Western analysis using antibodies against eIF1 or Gcd6 (as loading control). Two amounts of each extract differing by a factor of two were loaded in successive lanes. Signal intensities from four biological replicates were quantified and mean eIF1/Gcd6 ratios are listed below the blot with S.E.Ms. *p<0.05 (B) Strains from Figure 3B also harboring SUI1-lacZ (pPMB24) or SUI1-opt-lacZ (pPMB25) reporters, containing native or optimum context at positions −1 to −3, were assayed for β-galactosidase activities as in Figure 3D. Mean expression levels and S.E.M.s from four biological and two technical replicates are plotted, and ratio of mean expression levels of SUI1-lacZ reporters with optimized context to native context are listed below the histogram. *p<0.05 (C) β-galactosidase activities measured in WCEs of WT and uS7-D215L transformants harboring the el.uORF1 GCN4-lacZ reporters pC3502, pC4466, or pC3503 containing, respectively, the depicted optimum, weak, or poor context of uAUG-1; or the uORF-less GCN4-lacZ reporter pC3505 with mutated uAUG-1. Mean expression values with S.E.M.s were determined from three biological and two technical replicates and listed in columns 1 and 2. Cols. 3–4 gives the percentage of ribosomes translating the GCN4-lacZ ORF in the different constructs, calculated as a percentage of the GCN4-lacZ activity observed for the ‘no el. uORF1’ construct measured for the relevant construct shown in cols. 1–2. Cols. 5–6 gives the percentage of ribosomes translating el.uORF, calculated as 100% minus the percentage translating the GCN4-lacZ ORF shown in cols. 3–4. (*), p<0.05.
-
Figure 4—source data 1
Source data for Figure 4 and Figure 4—figure supplement 1.
Effects of Rps5-D215 substitutions on eIF1 expression, SUI1opt-lacZ: SUI1nat-lacZ expression ratios, and Rps5-D215L, -R225K, or -E144R on leaky scanning of el.uORF1 in GCN4-lacZ reporters.
- https://doi.org/10.7554/eLife.22572.007
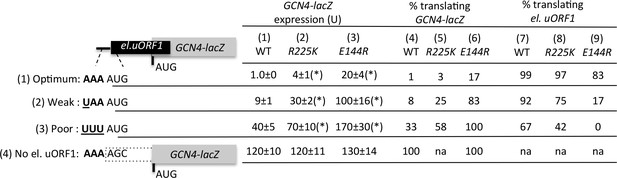
uS7 β-hairpin Ssu- substitutions R225K and E144R discriminate against AUG start codons in poor context.
The reporter data obtained previously for the uS7 β-hairpin Ssu- mutations R225K and E144R (Visweswaraiah et al., 2015) was analyzed as in (Figure 4C). Mean expression levels and S.E.M.s calculated from four biological and two technical replicates are presented. *p<0.05.
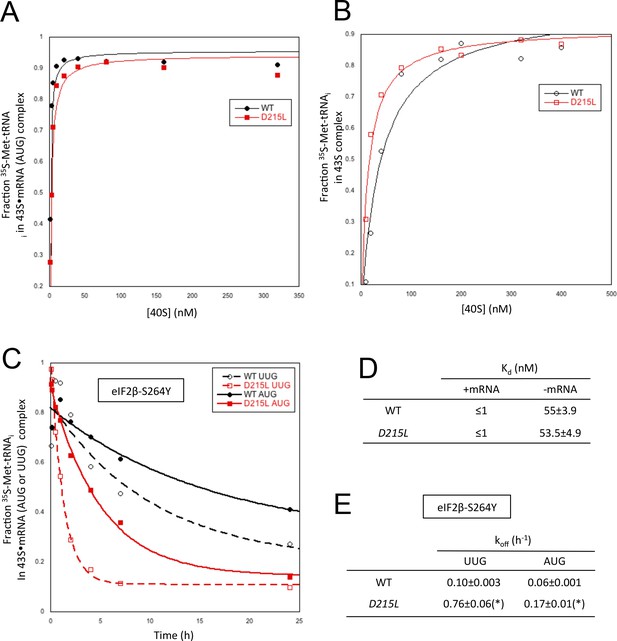
uS7 substitution D215L destabilizes PIN in vitro preferentially at UUG start codons.
(A, B) Determination of Kd values for TC with [35S]-Met-tRNAi binding to 40S·eIF1·eIF1A complexes assembled with WT or D215L mutant 40S subunits and either mRNA (AUG) (A) or without mRNA (B). (C) Analysis of TC dissociation kinetics from 43S·mRNA complexes assembled with WT or D215L mutant 40S subunits and mRNA(AUG) or mRNA(UUG), conducted using the eIF2β-S264Y Sui- variant of eIF2. Representative curves selected from three independent experiments are shown. (D, E) Kd and koff values with S.E.M.s from three independent experiments determined in (A–C). (*), p<0.05.
-
Figure 5—source data 1
Effects of Rps5-D215L on TC affinity for partial 43S and 43S·mRNA complexes, and rate of TC dissociation from partial 43S·mRNA complexes reconstituted with the eIF2β-S264Y variant of eIF2.
- https://doi.org/10.7554/eLife.22572.010
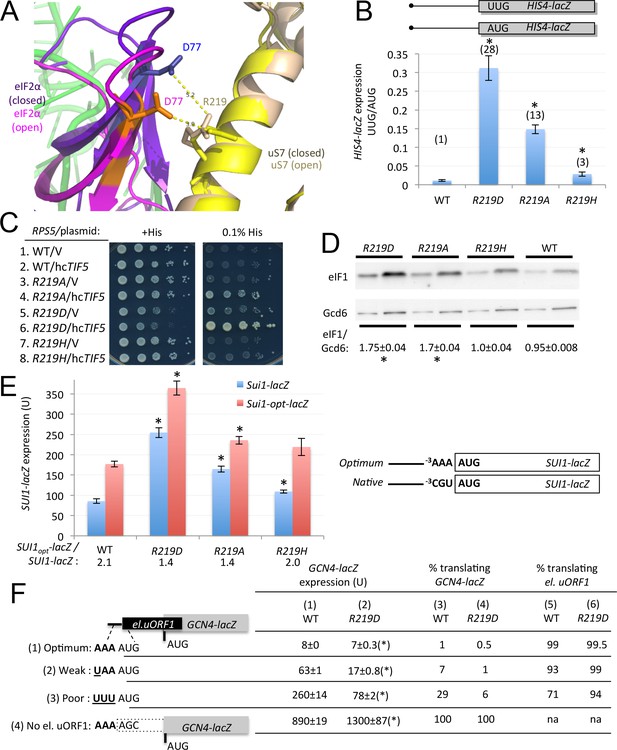
uS7 substitution R219D increases initiation at UUG codons and AUG codons in poor context.
(A) Overlay of py48S-open and py48S-closed showing uS7-R219/eIF2α-D77 interaction favored in the open complex (orange/yellow sticks). (B) Ratio of expression of HIS4-lacZ reporters with AUG or UUG start codons in transformants of JVY07 determined as in Figure 3D. Mean ratios and S.E.M.s calculated from four biological and two technical replicates. *p<0.05 (C) 10-fold serial dilutions of JVY07 transformants harboring the indicated RPS5 alleles and high-copy TIF5 plasmid p4438 or empty vector (V) spotted on SD+His+Ura (+His) or SD+Ura+0.0003 mM His (0.1% of usual His supplement; -His) and incubated at 30°C for 3d. (D) WCEs of three biological replicate strains from (B) subjected to Western analysis of eIF1 expression, as in Figure 4A. *p<0.05 (E) Expression of SUI1-lacZ or SUI1-opt-lacZ reporters in transformants of strains from (B), determined as in Figure 4B. Mean expression levels and S.E.M.s were calculated from four biological and two technical replicates. *p<0.05 (F) Expression of el.uORF1 GCN4-lacZ reporters in transformants of the WT or rps5-219D strains from (B), analyzed as in Figure 4C. (*), p<0.05.
-
Figure 6—source data 1
Source data for Figure 6 and Figure 6—figure supplement 1.
Effects of Rps5-R219 substitutions on HIS4-lacZ UUG:AUG expression ratios, eIF1 expression, SUI1opt-lacZ: SUI1nat-lacZ expression ratios, leaky scanning of el.uORF1 in GCN4-lacZ reporters, and polysome:monosome ratios.
- https://doi.org/10.7554/eLife.22572.012
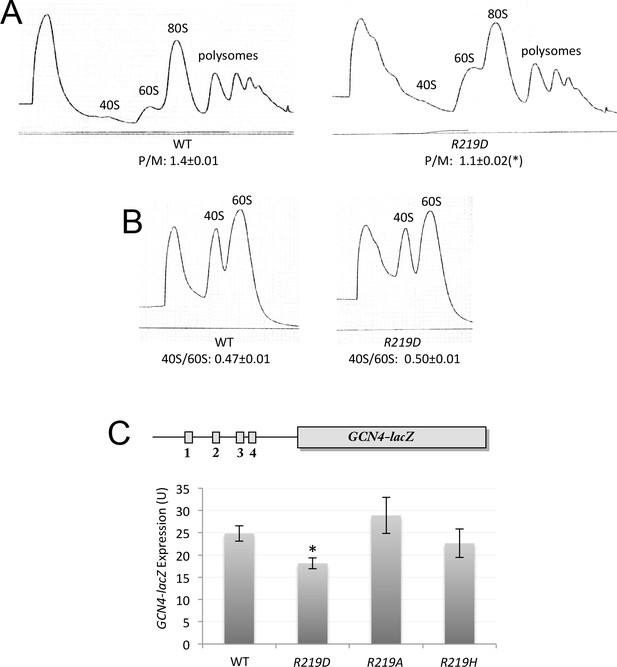
uS7 substitution R219D decreases bulk translation initiation but does not derepress translation of GCN4 mRNA.
(A–B) Polysome to monosome ratios (A) and 40S/60S ratios (B) in WT and rps5-R219D strains from Figure 6B, determined as in Figure 3E and Figure 3F, respectively, with mean ratios and S.E.M.s calculated from three biological replicates. (*), p<0.05 (C) Expression of the WT GCN4-lacZ reporter on plasmid p180 in transformants of strains from Figure 6B of the indicated RPS5 genotype, determined as in Figure 3D, with mean expression levels and S.E.M.s calculated from four biological and two technical replicates. *p<0.05.
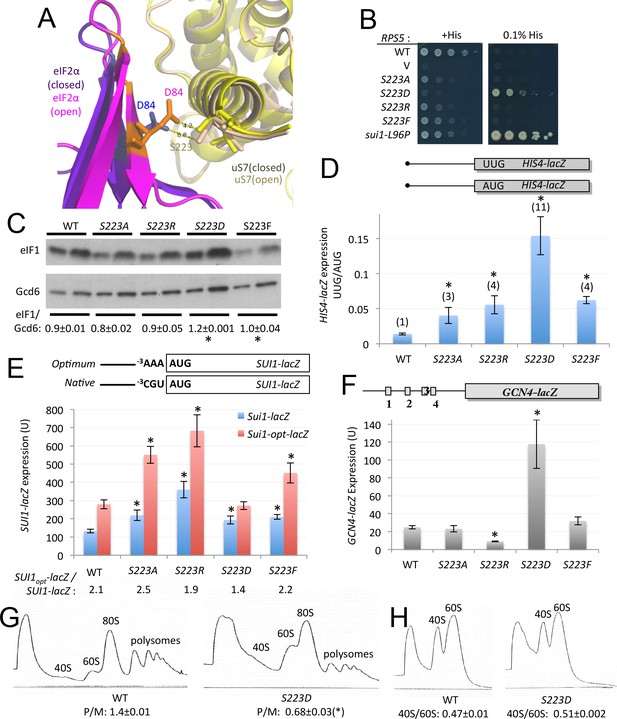
uS7 S223 substitutions decrease initiation fidelity in vivo.
(A) Overlay of py48S-open and py48S-closed complexes showing uS7-S223/eIF2α-D84 interaction favored in the open complex (orange/yellow sticks). (B) Dilutions of JVY07 transformed with the indicated RPS5 alleles and sui1-L96P strain H4564 spotted on SD+His+Ura+Trp (+His) or SD+Ura+Trp+0.0003 mM His (-His) and incubated at 30°C for 3 and 5 d, respectively. (C) WCEs of three biological replicate strains from (B) subjected to Western analysis of eIF1 expression as in Figure 4A. *p<0.05 (D) Ratio of expression of HIS4-lacZ reporters with AUG or UUG start codons in transformants of strains from (B), determined as described in Figure 3D. Mean ratios and S.E.M.s calculated from four biological and two technical replicates. *p<0.05 (E) Expression of SUI1-lacZ or SUI1-opt-lacZ reporters in transformants of strains from (B), determined as in Figure 4B. Mean expression levels and S.E.M.s calculated from four biological and two technical replicates. *p<0.05 (F) Expression of WT GCN4-lacZ in transformants of strains from (B), determined as in Figure 3D, with mean expression levels and S.E.M.s calculated from four biological and two technical replicates. *p<0.05 (G–H) Polysome to monosome ratios (G) and 40S/60S ratios (H) in WT and rps5-S223D strains from (B), determined as in Figure 3E–F with mean ratios and S.E.M.s calculated from three biological replicates. (*), p<0.05.
-
Figure 7—source data 1
Effects of Rps5-S223 substitutions on eIF1 expression, HIS4-lacZ UUG:AUG expression ratios, SUI1opt-lacZ: SUI1nat-lacZ expression ratios, GCN4-lacZ expression, and polysome:monosome ratios.
- https://doi.org/10.7554/eLife.22572.015
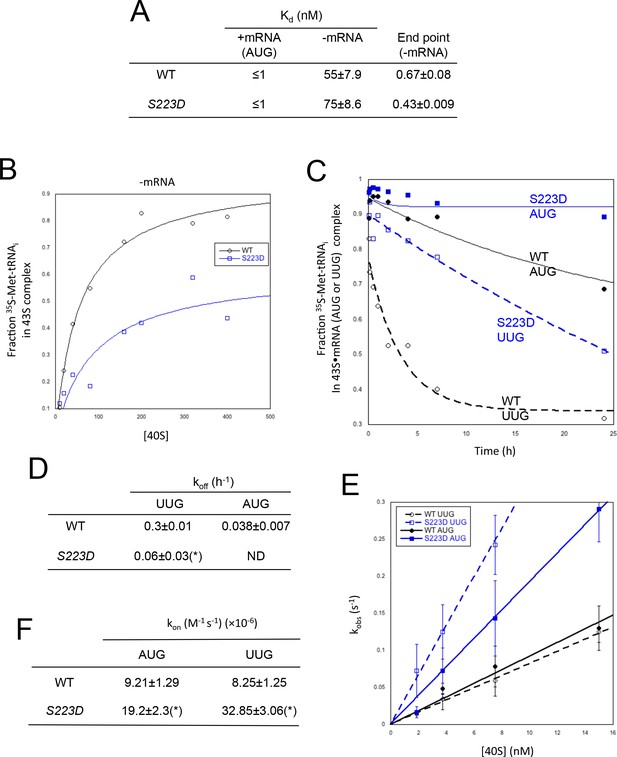
uS7 substitution S223D promotes PIN at UUG codons.
(A–B) Mean Kd and end-point values with S.E.M.s for binding of TC assembled with [35S]-Met-tRNAi to 40S·eIF1·eIF1A complexes reconstituted with WT or Rps5-S223D mutant 40S subunits and either mRNA (AUG) or without mRNA, determined from three independent experiments. A representative experiment is shown in (B). (C–D) Analysis of TC dissociation kinetics for 43S·mRNA complexes assembled with WT or Rps5-S223D mutant 40S subunits and either mRNA(AUG) or mRNA(UUG). A representative curve selected from three independent experiments is shown in (C), and mean koff values with S.E.M.s are given in (D). (*), p<0.05 (E–F) Determination of kon values for TC binding to 40S·eIF1·eIF1A complexes from plots of observed rate constants (kobs) vs 40S concentration for WT or Rps5-S223D mutant 40S subunits and mRNA(AUG or UUG) shown in (E) with S.E.M.s of kobs values for at least three independent experiments at each 40S concentration. Mean kon values with S.E.M.s calculated from three independent experiments are given in (F). (*), p<0.05.
-
Figure 8—source data 1
Effects of Rps5-S223D on TC affinity for partial 43S and 43S·mRNA complexes, and rates of TC association and dissociation from partial 43S·mRNA complexes.
- https://doi.org/10.7554/eLife.22572.017
Tables
Yeast strains employed in this study.
Strain | Genotype | Source or reference |
|---|---|---|
H4564 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) sui1Δ::hisG pPMB03 (sc LEU2 sui1-L96P) | |
JVY07 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 | |
JVY31 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 RPS5) | |
JVY76 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-D215L) | This study |
JVY77 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-D215A) | This study |
JVY78 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-D215F) | This study |
JVY85 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-R219D) | This study |
JVY86 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-R219A) | This study |
JVY87 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-R219H) | This study |
JVY91 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-S223A) | |
JVY92 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-S223D) | This study |
JVY93 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-S223R) | This study |
JVY94 | MATa ura3–52 trp1Δ−63 leu2–3,112 his4-301(ACG) kanMX6:PGAL1-RPS5 pJV09 (lc LEU2 rps5-S223F) | This study |
JVY11 | MATα ura3-∆0 leu2-∆0 his3∆−1 lys2-∆0 MET15 rps5∆::kanMX pJV38 (lc URA3 RPS5) | |
JVY15 | MATα ura3-∆0 leu2-∆0 his3∆−1 lys2-∆0 MET15 rps5∆::kanMX pJV13 (lc LEU2 RPS5) | |
JVY98 | MATα ura3-∆0 leu2-∆0 his3∆−1 lys2-∆0 MET15 rps5∆::kanMX pJV09 (lc LEU2 rps5-D215L) | This study |
JVY99 | MATα ura3-∆0 leu2-∆0 his3∆−1 lys2-∆0 MET15 rps5∆::kanMX pJV35 (lc LEU2 rps5-S223D) | This study |
Plasmids employed in this study.
Plasmid | Description* | Source or reference |
|---|---|---|
pJV09 | lc LEU2 RPS5 with BglII site engineered in pJV01 | |
pJV38 | lc URA3 RPS5 with BglII site engineered in pRS316 | |
pJV67 | lc LEU2 rps5-D215L | This study |
pJV68 | lc LEU2 rps5-D215A | This study |
pJV69 | lc LEU2 rps5-D215F | This study |
pJV76 | lc LEU2 rps5-R219D | This study |
pJV77 | lc LEU2 rps5-R219A | This study |
pJV78 | lc LEU2 rps5-R219H | This study |
pJV33 | lc LEU2 rps5-S223A | |
pJV82 | lc LEU2 rps5-S223R | This study |
pJV83 | lc LEU2 rps5-S223D | This study |
pJV84 | lc LEU2 rps5-S223F | This study |
p367 | sc URA3 HIS4(ATG)-lacZ | |
p391 | sc URA3 HIS4(TTG)-lacZ | |
p180 | sc URA3 GCN4-lacZ in YCp50 | |
p4280/YCpSUI3-S264Y-W | sc TRP1 SUI3-S264Y in YCplac22 | |
p4281/YCpTIF5-G31R-W | sc TRP1 TIF5-G31R in YCplac22 | |
p4438/YEplacTIF5-W | hc TRP1 TIF5 in YEplac112 | Christie Fekete |
pPMB24 | sc URA3 SUI1-lacZ | |
pPMB25 | sc URA3 SUI1-opt-lacZ | |
pC3502 | sc URA3 −3AAA−1 el.uORF1 GCN4-lacZ in YCp50 | |
pC4466 | sc URA3 −3UAA−1 el.uORF1 GCN4-lacZ in YCp50 | |
pC3503 | sc URA3 −3UUU−1 el.uORF1 GCN4-lacZ in YCp50 | |
pC3505 | sc URA3 el.uORF1-less GCN4-lacZ in YCp50 |
-
*lc, low copy number; sc, single copy; hc, high copy.





