Patterned cortical tension mediated by N-cadherin controls cell geometric order in the Drosophila eye
Figures
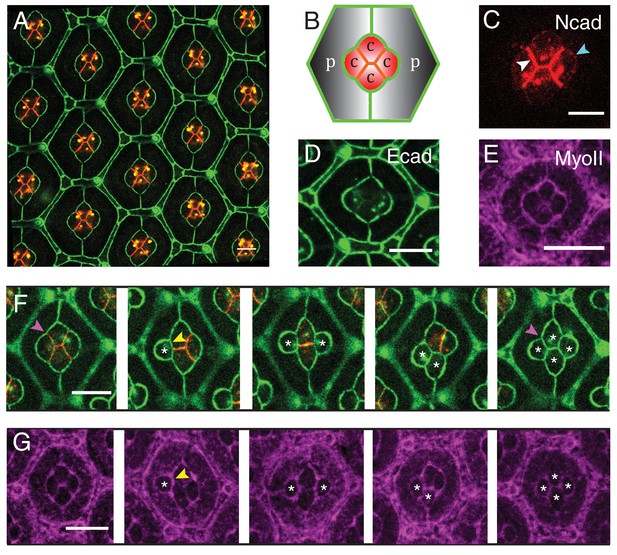
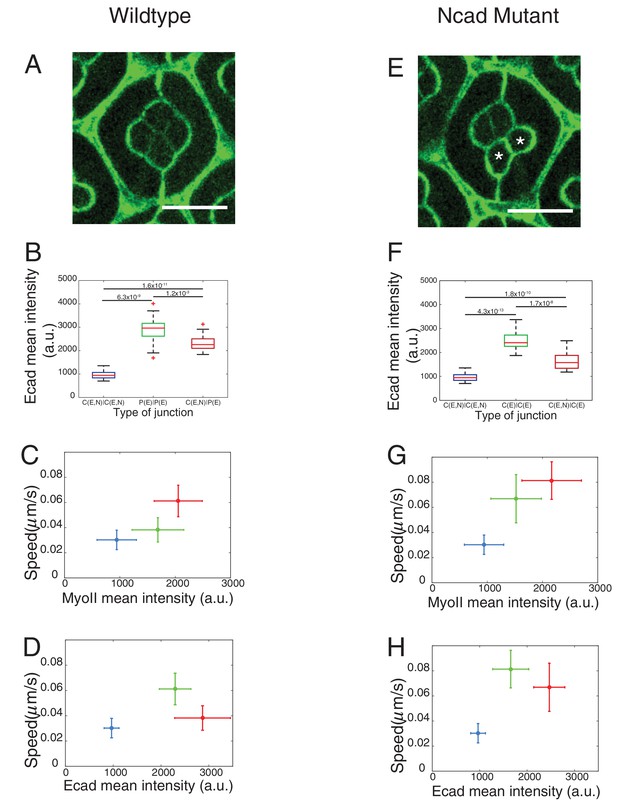
Patterns of Drosophila eye with the distributions of cadherins and Myosin-II (MyoII) in wildtype and NcadM19 mosaic ommatidia.
(A) Image of pupal retina at 41 hr after puparium formation (APF) consisting of repeating lattice structure called ommatidia labeled with Ecad::GFP (green) and Ncad::mKate2 (red). (B) A schematic of the most apical view of an ommatidium, which contains four cone cells (C) and two primary pigment cells (P), and the localization of cadherins (Ecad in green and Ncad in red). (C–E) An individual ommatidium with Ncad::GFP in red (C), Ecad::GFP in green (D), Zip::YFP in magenta (E). (F–G) Wildtype and NcadM19 mosaic ommatidia labelled with Ecad::GFP (green), Ncad (red) and Zip::YFP (magenta). NcadM19 cone cells are marked by white asterisks. Magenta arrowheads in (F) shows the angle change in full NcadM19 cone cells compared to wildtype. White arrowhead indicates the C|C contact with homophilic complexes and cyan arrowhead indicates the C|P contact in (C). Yellow arrowheads indicate one of the contacts at the interface between wildtype and NcadM19 cells to highlight the absence of Ncad adhesion in (F) and significant increase in MyoII levels in (G). Scale bar, 10 µm.
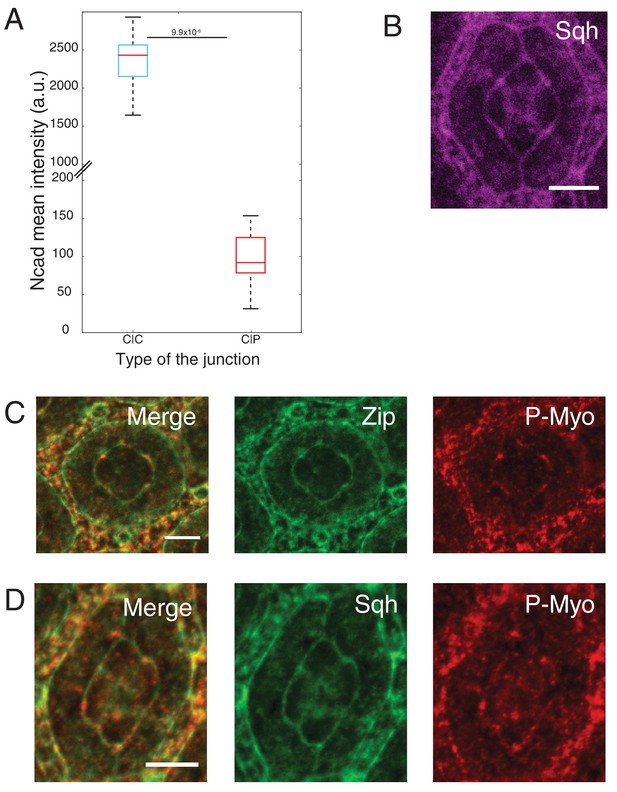
Ncad and MyoII levels using different reporters (Myosin-II light chain, Myosin-II heavy chain and Phospho-myosin-II light chain).
(A) Quantification of Ncad::GFP intensity in C|C (n = 12) and C|P (n = 20) contacts at 41 hr APF. P-value is shown above the black horizontal line (non-parametric Mann-Whitney U test). (B) An ommatidium with Sqh::GFP (magenta) in Sqh mutant background at 41 hr APF. (C) An ommatidium with Zip::YFP (green) co-stained with P-MyoII antibodies (red). (D) An ommatidium with Sqh::GFP (green) in Sqh mutant background co-stained with P-MyoII antibodies (red). Scale bar, 5 µm.
-
Figure 1—figure supplement 1—source data 1
Dataset for Figure 1- supplement figure 1A.
- https://doi.org/10.7554/eLife.22796.004
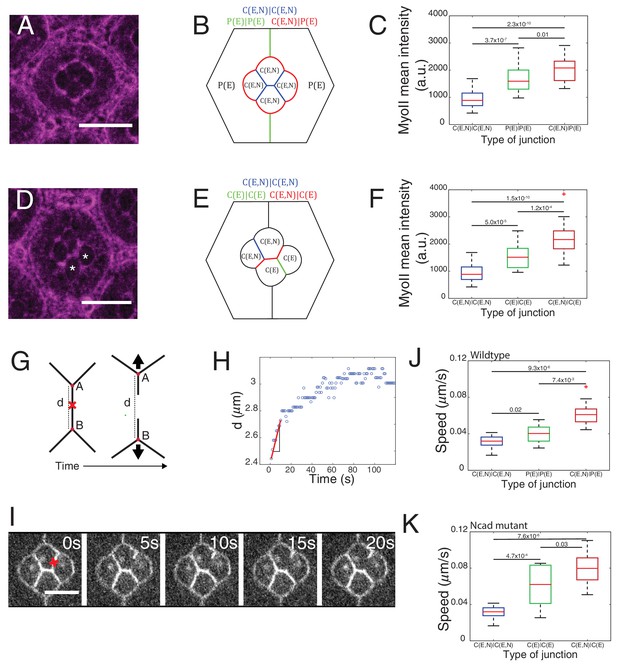
Differential MyoII levels and interfacial tensions at various cell contacts.
(A) Wildtype ommatidium with Zip::YFP represented by (B) a schematic that highlights three different types of contacts at cell interfaces that express Ecad or Ncad or both E and Ncad. C(E,N)|C(E,N) contact (blue) shared by two cone cells, P(E)|P(E) contact (green) shared by two primary pigment cells and C(E,N)|P(E) contact (red) shared by a cone and a primary pigment cell. Scale bar, 10 µm. (C) Quantification of MyoII intensity in C(E,N)|C(E,N) (n = 30), P(E)|P(E) (n = 22) and C(E,N)|P(E) (n = 36) contacts. P-values are shown above the black horizontal lines (non-parametric Mann-Whitney U test on pairs and Bonferroni correction). (D) A NcadM19 mosaic ommatidium with Zip::YFP. NcadM19 cells are marked by white asterisks. Scale bar, 10 µm. (E) A schematic represents the corresponding NcadM19 mosaic mutants highlighting C(E,N)|C(E,N) (blue), C(E)|C(E) (green) and C(E,N)|C(E) contacts (red). (F) Quantification of MyoII intensity in C(E,N)|C(E,N) (n = 30), C(E)|C(E) (n = 22) and C(E,N)|C(E) (n = 36) contacts. P-values are shown above the black horizontal lines. (G)-(K) Laser nanoablation experiments to estimate interfacial tension. (G) Schematic of a contact before (left) and after (right) ablation. Red cross represents the point of the ablation. Vertex A and B recoil changing distance 'd' after ablation. (H) Opening curve plots the distance’ d’ over time with a linear fit for initial time points to get the initial recoil speed. (I) Snapshot of an ablation at C(E,N)|C(E,N) contact in wildtype ommatidium, red cross indicates the ablation point. (J) Quantification of initial recoil speed of C(E,N)|C(E,N) (n = 14), P(E)|P(E) (n = 18) and C(E,N)|P(E) (n = 19) contacts in wildtype ommatidia. P-values are shown above the black horizontal lines. (K) Quantification of initial recoil speed in C(E,N)|C(E,N) (n = 14), C(E)|C(E) (n = 18) and C(E,N)|C(E) (n = 17) contacts in NcadM19 mosaic mutants. Scale bar, 5 µm. P-values are shown above the black horizontal lines.
-
Figure 2—source data 1
Dataset for Figure 2C,F,J and K.
- https://doi.org/10.7554/eLife.22796.006
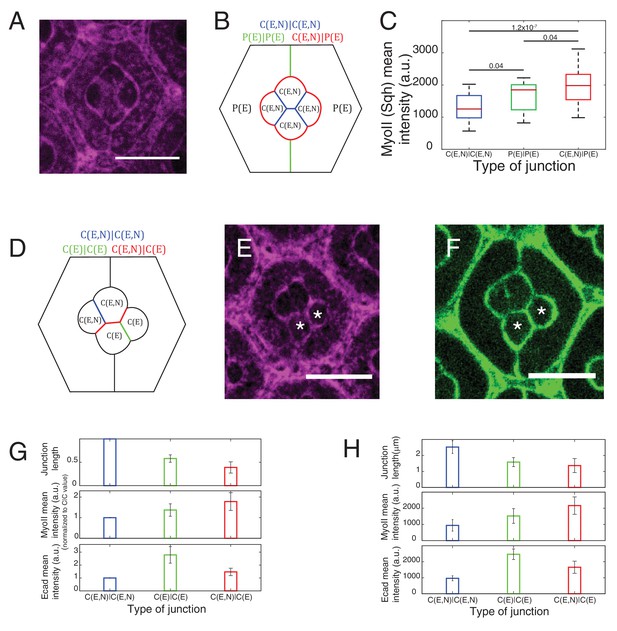
Junction length, cadherins and MyoII levels at different contacts.
(A) An ommatidium with Sqh::GFP in Sqh mutant background at 41 hr APF represented by (B) a schematic that highlights three different types of contacts at cell interfaces that express Ecad or Ncad or both Ecad and Ncad. C(E,N)|C(E,N) (blue), P(E)|P(E) (green) and C(E,N)|P(E) (red) contacts. Scale bar, 10 µm. (C) Quantification of MyoII intensity in C(E,N)|C(E,N) (n = 33), P(E)|P(E) (n = 21) and C(E,N)|P(E) (n = 50) contacts of ommatidia in (A). P-values are shown above the black horizontal lines. (D) A schematic of an ommatidium with 3 types of cell contacts in NcadM19 mosaic mutants highlighting C(E,N)|C(E,N) contact (blue), C(E)|C(E) contact (green) and C(E,N)|C(E) contact (red) (E–F) Image of MyoII (E) and Ecad (F) of NcadM19 mosaic ommatidium in (D). NcadM19 cells marked by white asterisks. (G) Normalized cell contact length (C|C (n = 15), C(E)|C(E) (n = 15), C(E,N)|C(E) (n = 29)), MyoII mean intensity (C(E,N)|C(E,N) (n = 10), C(E)|C(E) (n = 10), C(E,N)|C(E) (n = 20)) and Ecad mean intensity C(E,N)|C(E,N) (n = 15), C(E)|C(E) (n = 15), C(E,N)|C(E) (n = 29)) of the NcadM19 mosaic ommatidia in (D). (H) Cell contact length C(E,N)|C(E,N) (n = 38), C(E)|C(E) (n = 36), C(E,N)|C(E) (n = 26)) MyoII mean intensity (same as Figure 2F) and Ecad mean intensity in NcadM19 mosaic ommatidia. (same as Figure 5—figure supplement 3F).
-
Figure 2—figure supplement 1—source data 1
Dataset for Figure 2—figure supplement 1C,G and H.
- https://doi.org/10.7554/eLife.22796.008
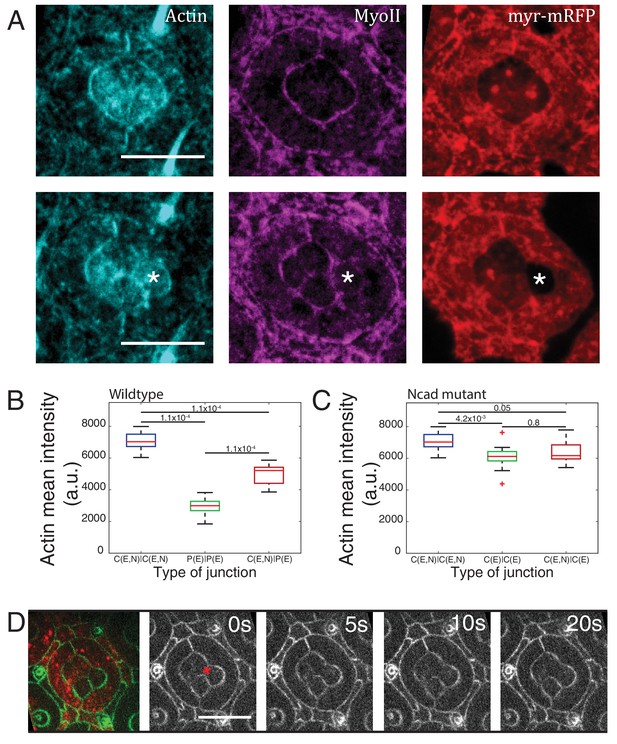
NcadM19 mosaic ommatidium interfacial tension measurements and F-actin distribution.
(A) Wildtype (upper panels) and NcadM19 mosaic ommatidia (lower panels) with phalloidin staining for F-actin in cyan, Zip::YFP in magenta and wildtype cells are marked in myr-RFP (red) for locating the cell contacts. (B) Quantification of F-actin intensity in wildtype C|C (n = 12), P(E)|P(E) (n = 12) and C(E,N)|P(E) (n = 12) contacts. P-values are shown above the black horizontal lines. (C) Quantification of F-actin intensity in NcadM19 mosaic ommatidia C(E,N)|C(E,N) (n = 12), C(E)|C(E) (n = 15) and C(E,N)|C(E) (n = 15) contacts. Scale bar, 10 µm. P-values are shown above the black horizontal lines. (D) Image of a NcadM19 mosaic ommatidium with Ecad::GFP (green) and wildtype cells in red, followed by snapshot of an ablation at a C(E,N)|C(E) contact (red cross). Scale bar, 10 µm.
-
Figure 2—figure supplement 2—source data 1
Dataset for Figure 2—figure supplement 2B and C.
- https://doi.org/10.7554/eLife.22796.010
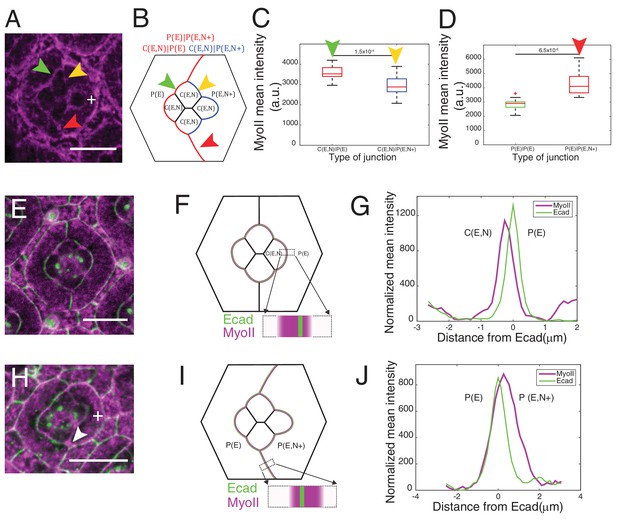
Misexpression of Ncad in primary pigment cells and MyoII accumulation and MyoII asymmetry at cell contacts.
(A) An ommatidium with Ncad misexpressed in one of the primary pigment cells (white +) with Zip::YFP in magenta. Green arrowhead indicates the C(E,N)|P(E) contact. Yellow and red arrowheads indicate the modified C(E,N)|P(E,N+) and P(E)|P(E,N+) contacts respectively. (B) A schematic of Ncad misexpression ommatidium with the modified C(E,N)|P(E,N+) (blue), wildtype C(E,N)|P(E) and modified P(E)|P(E,N+) (red) contacts. (C) Quantification of MyoII intensity in C(E,N)|P(E) (n = 20) and C(E,N)|P(E,N+) (n = 20) contacts. P-value is shown above the black horizontal line. (D) Quantification of MyoII intensity in P(E)|P(E) (n = 16) and P(E)|P(E,N+) (n = 16) contacts. P-value is shown above the black horizontal line. (E) Wildtype ommatidium with Ecad::GFP (green) and Sqh::Ch (magenta). (F) Schematic with a zoom-in of a C(E,N)|P(E) contact shared by cone cell and primary pigment cell representing the asymmetric distribution of MyoII. (G) Average linescan of Sqh::Ch (magenta) intensity with respect to Ecad::GFP intensity (green) normal to interfaces (n = 10 interfaces). (H) An ommatidium with Ncad misexpressed in one of the primary pigment cell (white +) with Sqh::Ch (magenta). White arrowhead indicates the modified P(E)|P(E,N+) contact. (I) Schematics with a zoom-in of a modified P(E)|P(E,N+) contact shared by primary pigment cell and Ncad misexpressed primary pigment cell representing the asymmetric distribution of MyoII. (J) Average linescan of Sqh::Ch intensity (magenta) with respect to Ecad::GFP intensity (green) (n = 13 interfaces). Scale bar 10 µm.
-
Figure 3—source data 1
Dataset for Figure 3C,D,G and H.
- https://doi.org/10.7554/eLife.22796.014
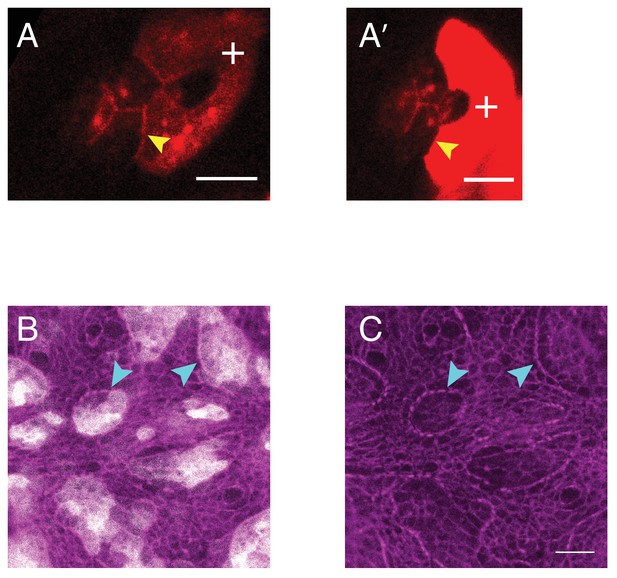
Misexpression of Ncad in primary pigment cell in retinas and larval wing pouch.
(A–A’) Examples of ommatidium misexpressing Ncad in one of the primary pigment cells (white +) with antibody staining for Ncad in red. Yellow arrowhead indicates the C(E,N)|P(E,N+) cell contact with homophilic Ncad. Scale bar, 5 µm. (B–C) Larval wing pouch with Sqh::Ch (magenta) in (C) and misexpression of Ncad clones (grey in (B). Accumulation of MyoII at clonal boundaries indicated by cyan arrowheads. Scale bar, 10 µm.
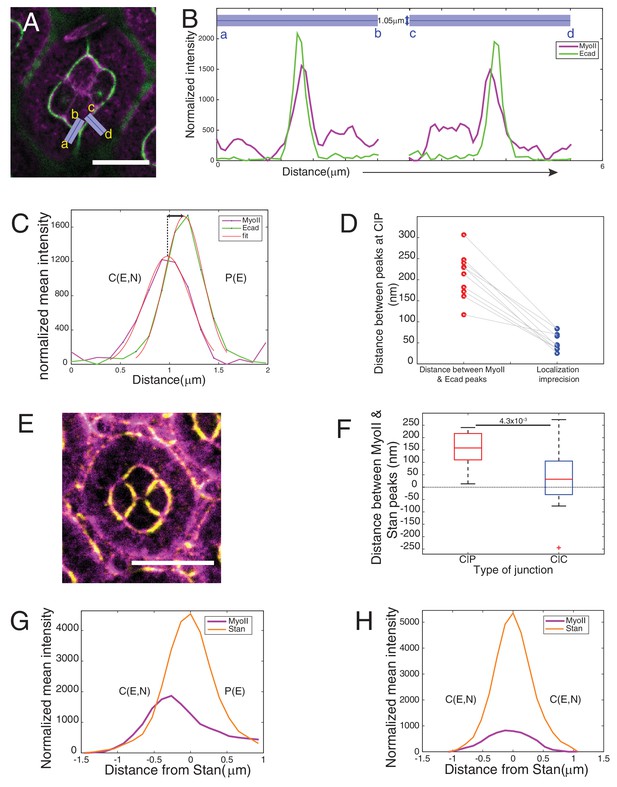
Asymmetry of MyoII localization at different contacts.
(A–B) Individual linescan of Sqh::Ch (magenta) and Ecad::GFP (green) from the purple lines (linewidth 1.05 mm) drawn orthogonal to cell contacts from cone cell to primary pigment cell in a wildtype ommatidium in (A). (C) Individual linescan orthogonal to a C(E,N)|P(E) contact. Peak positions were determined by fitting intensity profiles with Gaussian. (D) Distance between Ecad and MyoII peak positions at C(E,N)|P(E) contacts and the respective imprecision in localization of the two peaks. (E) Wildtype ommatidium with Zip::YFP in magenta and antibody staining for Starry night (Stan) (yellow). (F) Distance between Ecad and MyoII peak positions at C(E,N)|P(E) and C(E,N)|C(E,N) contacts obtained from individual linescan (n = 15 different contacts for each contact type). P-value is shown above the black horizontal line. (G) Average linescan of Zip::YFP (magenta) with respect to Stan (orange) at C(E,N)|P(E) contact (n = 15 contacts). (H) Average linescan of Zip::YFP (magenta) with respect to Stan (orange) at C(E,N)|C(E,N) (n = 15 contacts). Scale bar 10 µm.
-
Figure 3—figure supplement 2—source data 1
Dataset for Figure 3—figure supplement 2D,F,G and J.
- https://doi.org/10.7554/eLife.22796.017
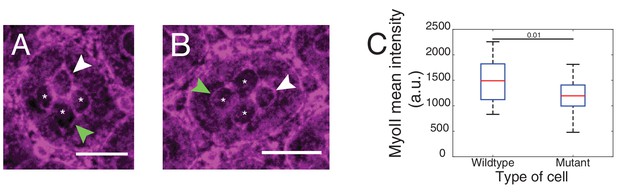
MyoII levels of a single wildtype cone cell in NcadM19 mosaic ommatidium.
(A) MyoII (Zip::YFP) (in magenta) in wildtype equatorial cone cell expressing both Ecad and Ncad (indicated by white arrowhead) surrounded by NcadM19 cone cells. (B) MyoII in wildtype posterior expressing both Ecad and Ncad (indicated by white arrowhead) surrounded by NcadM19 cone cells. (C) Quantification of MyoII intensity in contour of 1(E,N)|2(E) contacts of wildtype cone cell (n = 41, white arrowhead in images (A), (B) and in contour of 1(E)|2(E) NcadM19 cone cells (n = 41, green arrowhead in images (A), (B)). P-value is shown above the black horizontal line.
-
Figure 3—figure supplement 3—source data 1
Dataset for Figure 3—figure supplement 3C.
- https://doi.org/10.7554/eLife.22796.019
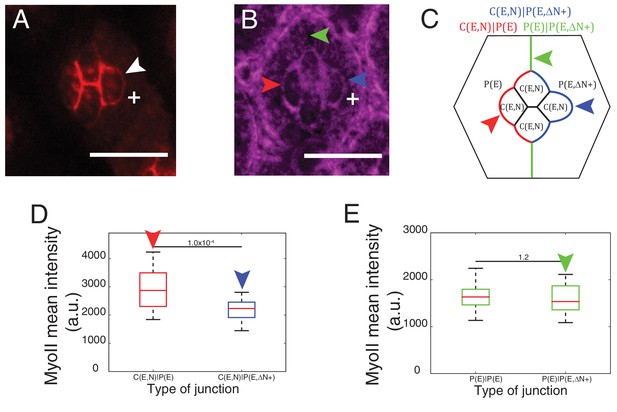
Cytoplasmic part of Ncad is required for MyoII accumulation in heterotypic contacts.
(A–B) An ommatidium misexpressing extracellular part of Ncad in one of the primary pigment cells (white +) with Ncad (A) and Zip::YFP (B). White arrowhead indicates the C(E,N)|P(E,ΔN+) cell contact with homophilic Ncad in (A), red arrowhead indicates C(E,N)|P(E,ΔN+) wildtype cell contact, blue arrowhead indicates modified C(E,N)|P(E) cell contact and green arrowhead indicates the unchanged P(E)|P(E,ΔN+) cell contact. (C) Schematic of ommatidium misexpressing extracellular part of Ncad shows the modified cell contacts, C(E,N)|P(E) contact (blue), wildtype C(E,N)|P(E) contact (red) and unaffected P(E)|P(E,ΔN+) contact (green). (D) Quantification of MyoII intensity in C(E,N)|P(E) (n = 28) and C(E,N)|P(E,ΔN+) (n = 28). P-value is shown above the black horizontal line. (E) Quantification of MyoII intensity in wildtype P(E)|P(E) (n = 19) and unaffected P(E)|P(E,ΔN+) contact (n = 19). Scale bar, 10 µm. P-value is shown above the black horizontal line.
-
Figure 4—source data 1
Dataset for Figure 4D and E.
- https://doi.org/10.7554/eLife.22796.021
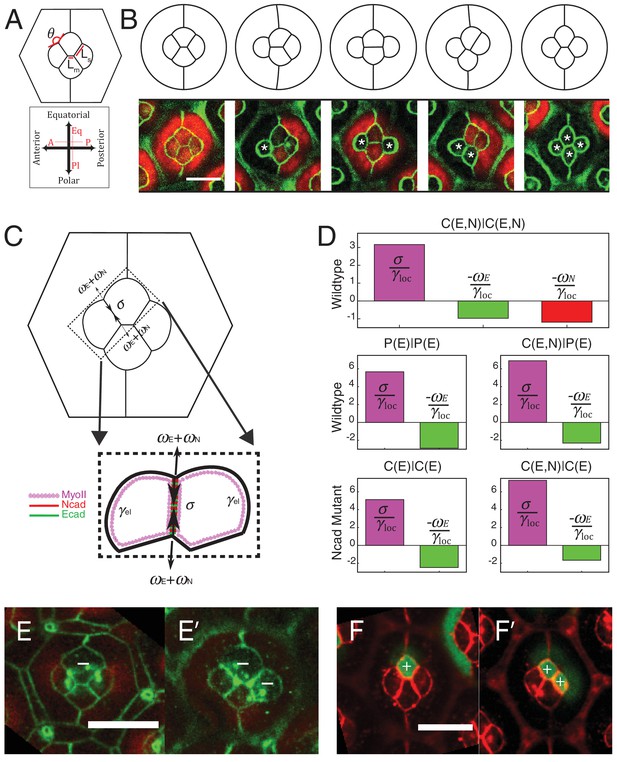
Simulations of cone cell shapes and contribution of cadherins and MyoII to cell shapes.
(A) Schematics of two axes of polarity, A-P and Eq-Pl, of cone cells (bottom) and fit parameters measured in experiments and simulations, contact angle between cone cell and primary pigment cell (θ), ratio of contact length shared by A/P and Eq/Pl cell (Ls) to contact length shared by Eq and Pl cells (Lm) (top). (B) Comparison of experimental images (lower panel) to the simulations (upper panel), NcadM19 cells are marked by white asterisks. (C) Schematic of force balance resulting from adhesion of Ecad (ωE, green) and Ncad (ωN, red), MyoII dependent cortical tension at the cell contact (σ) and cortex elasticity due to actomyosin at the cell perimeter (γel) (both in magenta). (D) Relative contribution of MyoII dependent cortical tension (σ), Ecad adhesion (wE) and Ncad adhesion (wN) to the local tension term γloc for all contact types in wildtype and NcadM19 mosaic mutants. (E–E') Image of the ommatidium with (E) Eq and (E') Eq and Pl cone cells SqhAx3 mutant (white -). β-catenin staining in green. (F–F') Image of the ommatidium with (F) Eq and (F') Eq and Pl cone cells expressing constitutively active form of Sqh, SqhT20ES20E (white +), β-catenin staining in red. Scale bar, 10 µm.
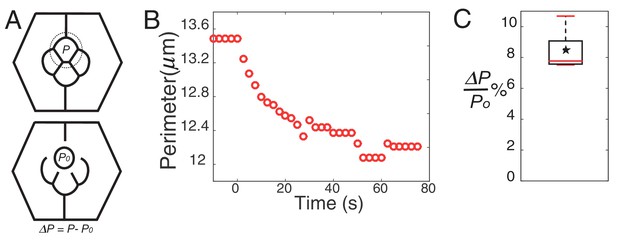
Perimeter elasticity and determination of elastic constant (K).
(A) Schematic of a circle ablation experiment. Dotted circle marks the ablation position with P, perimeter before ablation and P0, perimeter after ablation with ΔP= P-P0. (B) Plot of a perimeter change over the time from an experiment. (C) Boxplot showing percentage change of perimeter (n = 7).
-
Figure 5—figure supplement 1—source data 1
Dataset for Figure 5—figure supplement 1C
- https://doi.org/10.7554/eLife.22796.024
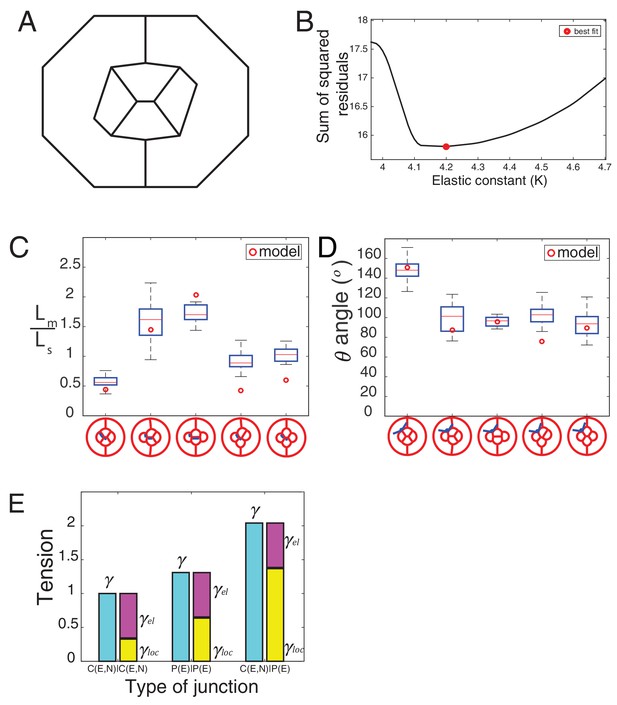
Elastic and local tension contribution to interfacial tension and comparison of simulation to experiment.
(A) Initial configuration used in Surface Evolver before the minimization of the energy function. (B) Plot of the sum of squared residuals as a function of K. The best fit is obtained for K = 4.2 (red circle). (C) Graph of fit-parameter Lm/Ls (n1 = 43, n2 = 9, n3 = 8, n4 = 28, n5 = 15) for experimental images in wildtype and NcadM19 mosaic configurations, and comparison to the respective simulation value. (D) Graph of fit-parameter angle (θ) (n1 = 210, n2 = 9, n3 = 8, n4 = 28, n5 = 64) for experimental images in wildtype and NcadM19 mosaic configurations, and comparison to the respective simulation value. (E) Tensions γ, γel, γloc for the three different types of wildtype contacts.
-
Figure 5—figure supplement 2—source data 1
Dataset for Figure 5—figure supplement 2C and D.
- https://doi.org/10.7554/eLife.22796.026
Ecad intensity measurements and correlation of interfacial tension to molecular distributions.
(A) Wildtype ommatidium with Ecad::GFP. (B) Quantification of Ecad intensity in wildtype ommatidia with C(E,N)|C(E,N) (n = 38), P(E)|P(E) (n = 18) and C(E,N)|P(E) (n = 28) contacts. P-values are shown above the black horizontal lines. (C–D) Correlation of interfacial tension to the mean intensity of MyoII (C) and Ecad (D) at three different cell contacts in wildtype ommatidia. Correlation of interfacial tension to MyoII mean intensity at three different cell contacts in (E) A NcadM19 mosaic mutant ommatidium with Ecad::GFP. NcadM19 marked by white asterisks. (F) Quantification of Ecad intensity in C|C (n = 38), C(E)|C(E) (n = 36) and C(E,N)|C(E) (n = 26) contacts. P-values are shown above the black horizontal lines. (G–H) Correlation of interfacial tension to the mean intensity of MyoII (G) and Ecad (H) at three different cell contacts in wildtype ommatidia. Scale bar, 10 µm.
-
Figure 5—figure supplement 3—source data 1
Dataset for Figure 5—figure supplement 3B and F.
- https://doi.org/10.7554/eLife.22796.028
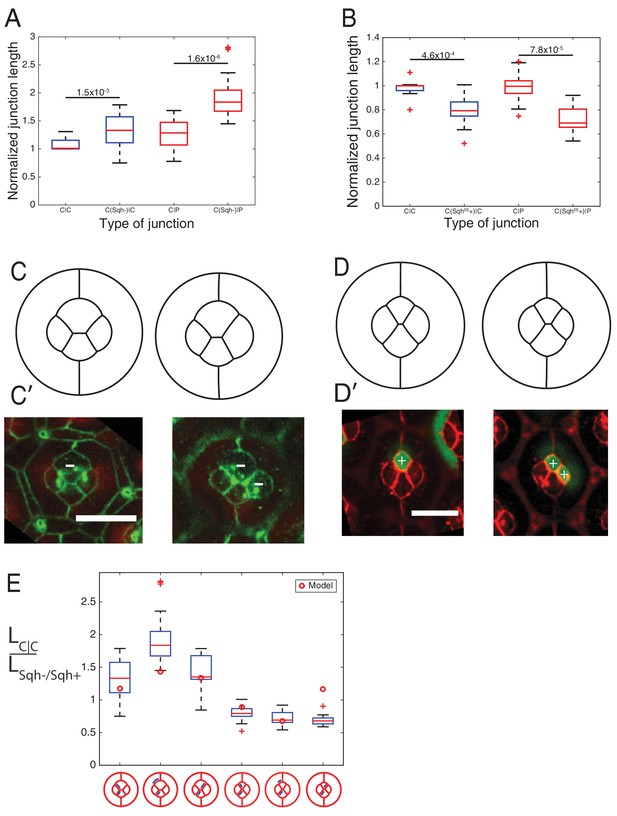
MyoII perturbations and simulations.
(A) Comparison of junction length of wildtype C|P contact, modified contacts containing one SqhAx3 mutant cell (C(Sqh-)|C or C(Sqh-)|P) to wildtype C|C contacts in experiments. P-values are shown above the black horizontal lines. (B) Comparison of junction length between wildtype C|P contacts(C|C or C|P), and modified contacts misexpressed active SqhT20ES20E (SqhEE) in one cell (C(SqhEE+)|C or C(SqhEE+)|P) to wildtype C|C contacts in experiments. P-values are shown above the black horizontal lines. (C–C′) Comparison of simulations (C) to the experimental images (C′) of SqhAx3 mutant (white -), β-catenin staining in green. (D–D’) Comparison of simulations (D) to the experimental images (D′) of SqhEE misexpression (white +), β-catenin staining in red. Scale bar, 10 µm. (E) Ratio of contact length in C|C to different modified contacts with SqhAx3 or SqhEE cells.
-
Figure 5—figure supplement 4—source data 1
Dataset for Figure 5—figure supplement 4A,B and E.
- https://doi.org/10.7554/eLife.22796.030
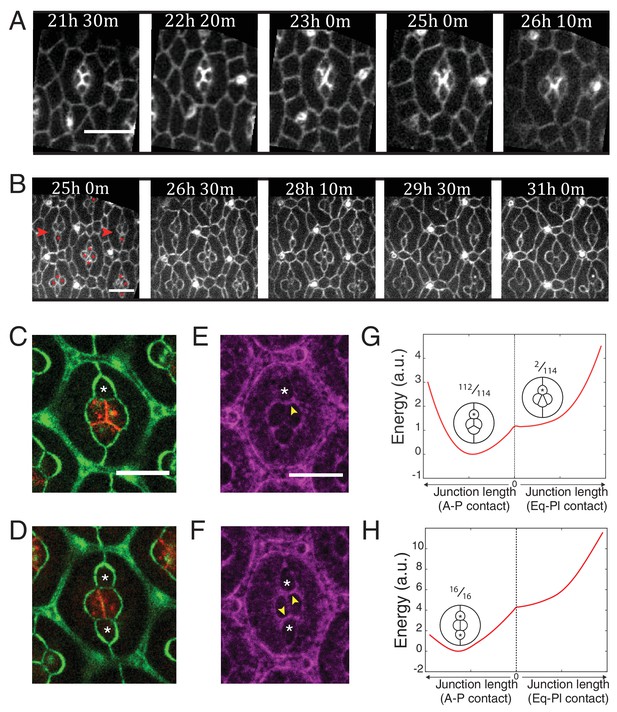
Ncad mediated MyoII contractility impacts on cone cell arrangements.
(A) Snapshots of a movie at different APF from wildtype retina labelled with β-cat::GFP. Scale bar, 5 µm. (B) Snapshots of a movie at different APF from NcadM19 mosaic mutant with Ecad::GFP and NcadM19 cells (red asterisks). Mosaic ommatidia that failed to undergo normal cell rearrangement are indicated by red arrowheads. Scale bar, 5 µm. (C), (E) Equatorial NcadM19 cone cell (white asterisk) in mosaic mutant with Ecad::GFP (green) and Ncad (red) in (C) and Zip::YFP (magenta) in (E) (both (C) and (E) total n = 112). (D), (F) Image of equatorial and polar NcadM19 cone cells (white asterisks) with Ecad::GFP (green) and Ncad (red) in (D) and Zip::YFP (magenta) in (F) (both (D) and (F) total n = 16). (G) Energy profile of ommatidia with an equatorial NcadM19 cone cell as a function of the central contact length (left direction: vertical contact length, right direction: horizontal contact length). Diagrams show corresponding simulations, with occurrence numbers observed experimentally. (H) Energy profile of ommatidia with equatorial and polar NcadM19 cone cells as a function of the central contact length (left direction: vertical contact length, right direction: horizontal contact length). Diagrams show corresponding simulations, with occurrence numbers observed experimentally.
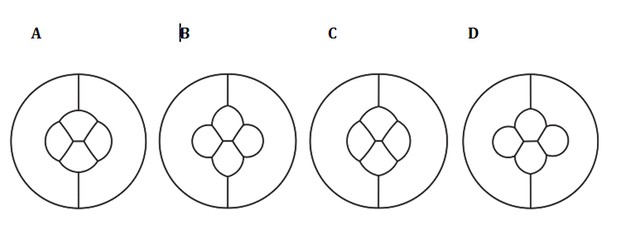
Simulation of an ommatidium with (A) differential adhesion and differential contraction (wildtype), (B) Uniform adhesion and differential contraction, (C) Differential adhesion and uniform contraction (D) Uniform adhesion and uniform contraction.
Videos
Laser nano-ablation of C(E,N)|C(E,N) contact in wildtype ommatidium.
Ablation at 00:00:00. Frame rate is 1 s/frame. Labelling: β-cat::GFP. Scale bar, 5 μm.
Laser nano-ablation of C(E,N)|C(E) contact in NcadM19 mosaic ommatidium with polar (Pl) and posterior (P) cone cells(see Figure 5A for cone cell axes of polarity) lacking Ncad.
Ablation at 00:00:00. Frame rate is 250 ms/frame. Labelling: Ecad::GFP. Scale bar, 5 μm.
Laser nano-ablation of target perimeter measurement (Δp/po).
Ablation at 00:00. Frame rate is 250 ms/frame. Labelling: β-cat::GFP. Scale bar, 5 μm.
A-P to Eq-Pl transition in wildtype retina.
Movies starting from 21:30:00 APF. Frame rate is 10 min/frame. Labelling: β-cat::GFP. Scale bar, 5 μm.
Defects in cell rearrangements in NcadM19 mosaic mutants.
Movies starting from 25:00:00 APF. Frame rate is 10 min/frame. Labelling: Ecad::GFP. Scale bar, 5 μm.
Additional files
-
Supplementary file 1
Table 1: MyoII levels in different experiments at different contact types. Table 2: Junction length, MyoII level, Ecad level and Ncad level in different experiments at different contact types. Table 3: Statistical value for all quantifications. Table 4: Oligos used in generating CRISPR/Cas9 mediated knock-in Ncad::mKate2 flies.
- https://doi.org/10.7554/eLife.22796.035





