Role of D-aminoacyl-tRNA deacylase beyond chiral proofreading as a cellular defense against glycine mischarging by AlaRS
Figures
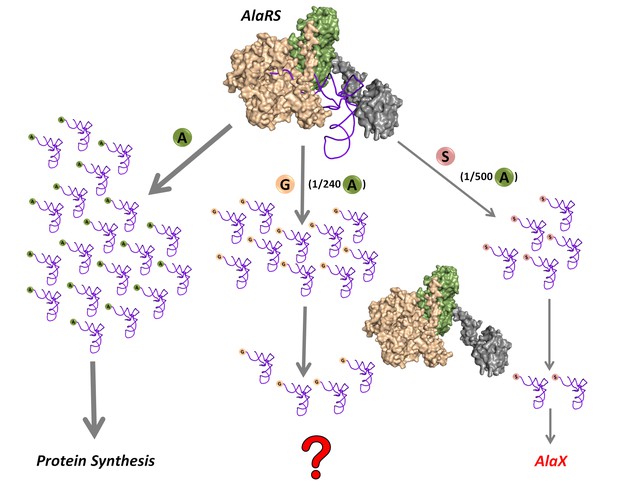
Mischarging by AlaRS.
AlaRS activates and charges alanine (A) to form cognate Ala-tRNAAla which is routed for protein synthesis. In this process, AlaRS also misactivates glycine (G) and serine (S) at frequencies of 1 per 240 alanine and 1 per 500 alanine, respectively (Tsui and Fersht, 1981). The two non-cognate amino acids are then charged on tRNAAla to produce Gly-tRNAAla and Ser-tRNAAla species, with glycine mischarging being nearly twice that of serine. Since AlaRS does not distinguish much between Gly-tRNAAla and Ser-tRNAAla while clearing the two, higher levels of Gly-tRNAAla might accumulate in the cell. However, there are additional free-standing trans-editing factors called AlaX (found in all domains of life but not in all organisms), which are known to edit mainly Ser-tRNAAla. This leads to a fundamental question as to how the problem of Gly-tRNAAla editing is solved in the cellular context.
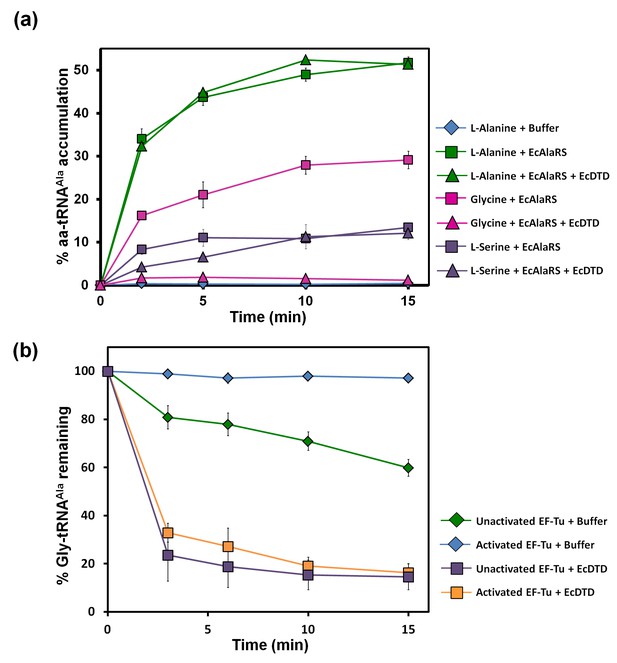
Misacylation of tRNAAla with glycine by AlaRS and its prevention/rectification by DTD.
(a) Aminoacylation of tRNAAla by EcAlaRS in the presence of activated EF-Tu: L-alanine (green square), L-alanine and 10 pM EcDTD (green triangle), glycine (pink square), glycine and 10 pM EcDTD (pink triangle), L-serine (purple square), L-serine and 10 pM EcDTD (purple triangle). No enzyme control (blue diamonds) reaction had all the components of the reaction (with L-alanine) except for EcAlaRS. (b) Deacylation of Gly-tRNAAla in the presence of unactivated EF-Tu (green diamond), activated EF-Tu (blue diamond), 5 pM EcDTD and unactivated EF-Tu (purple square), 5 pM EcDTD and activated EF-Tu (orange square). Error bars indicate one standard deviation from the mean of triplicate readings.
-
Figure 2—source data 1
Misacylation of tRNAAla and deacylation of Gly-tRNAAla in the presence of EF-Tu.
- https://doi.org/10.7554/eLife.24001.005
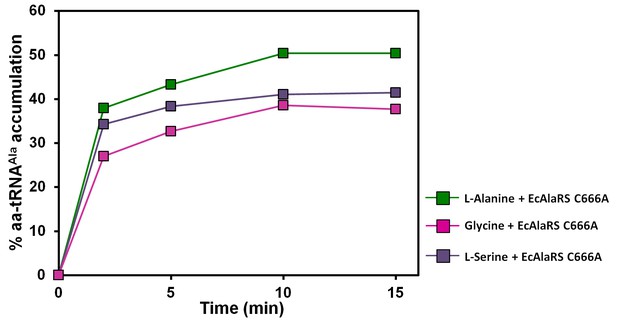
Accumulation of Ala/Gly/Ser-tRNAAla during aminoacylation by EcAlaRS C666A in the presence of EF-Tu.
Aminoacylation of tRNAAla by EcAlaRS C666A in the presence of activated EF-Tu: L-alanine (green square), glycine (pink square), L-serine (purple square).
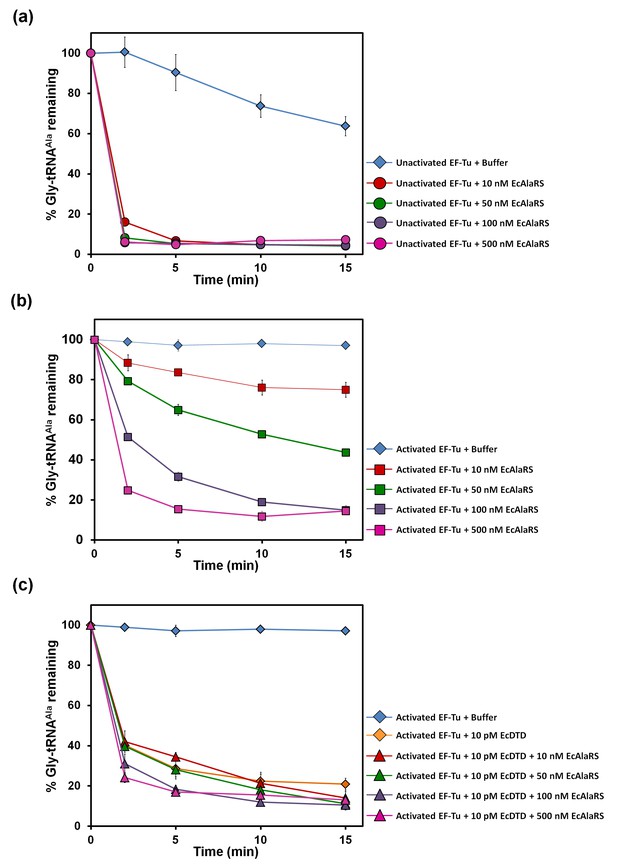
DTD has higher activity than AlaRS for the editing of Gly-tRNAAla.
(a) Deacylation of Gly-tRNAAla in the presence of unactivated EF-Tu: buffer (blue diamond), 10 nM EcAlaRS (red circle), 50 nM EcAlaRS (green circle), 100 nM EcAlaRS (purple circle), 500 nM EcAlaRS (pink circle). (b) Deacylation of Gly-tRNAAla in the presence of activated EF-Tu: buffer (blue diamond), 10 nM EcAlaRS (red square), 50 nM EcAlaRS (green square), 100 nM EcAlaRS (purple square), 500 nM EcAlaRS (pink square). (c) Deacylation of Gly-tRNAAla by EcDTD and increasing concentration of EcAlaRS: buffer (blue diamond), 10 pM EcDTD (orange diamond), 10 pM EcDTD and 10 nM EcAlaRS (red triangle), 10 pM EcDTD and 50 nM EcAlaRS (green triangle), 10 pM EcDTD and 100 nM EcAlaRS (purple triangle), 10 pM EcDTD and 500 nM EcAlaRS (pink triangle). Error bars indicate one standard deviation from the mean of triplicate readings.
-
Figure 3—source data 1
Deacylation of Gly-tRNAAla in the presence of unactivated and activated EF-Tu.
- https://doi.org/10.7554/eLife.24001.008
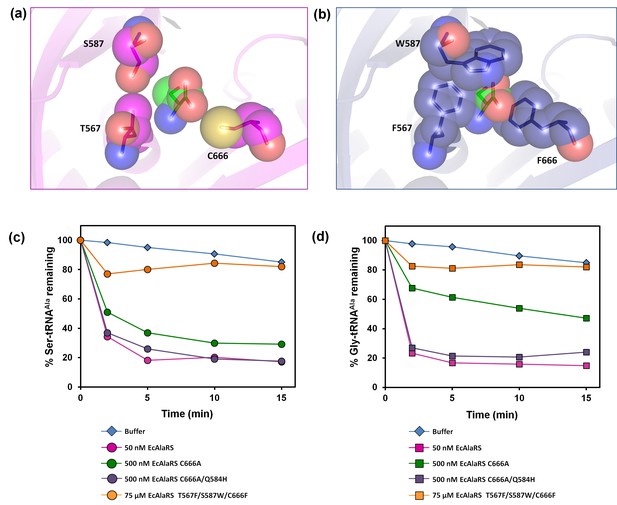
E. coli AlaRS editing site mutants.
Homology model of E. coli AlaRS depicting serine (green sticks/spheres) in the editing site. E. coli AlaRS cis-editing domain was modeled using A. fulgidus AlaRS (PDB id: 2ZTG) as a template, whereas the position and orientation of serine in the model corresponds to that observed in serine-bound P. horikoshii AlaX structure (PDB id: 1WNU). (a) In the wild-type enzyme, residues selected for mutagenesis are represented as megenta sticks/spheres, showing an open pocket for substrate binding. (b) In AlaRS T567F/S587W/C666F, the mutated bulkier residues are depicted as blue sticks/spheres, showing occlusion of the pocket to prevent substrate binding. (c) Deacylation of Ser-tRNAAla by buffer (blue diamond), 50 nM EcAlaRS (pink circle), 500 nM EcAlaRS C666A (green circle), 500 nM EcAlaRS C666A/Q584H (purple circle), 75 µM EcAlaRS T567F/S587W/C666F (orange circle). (d) Deacylation of Gly-tRNAAla by buffer (blue diamond), 50 nM EcAlaRS (pink square), 500 nM EcAlaRS C666A (green square), 500 nM EcAlaRS C666A/Q584H (purple square), 75 µM EcAlaRS T567F/S587W/C666F (orange square).
-
Figure 4—source data 1
Deacylation of Ser-tRNAAla and Gly-tRNAAla by E. coli AlaRS editing site mutants.
- https://doi.org/10.7554/eLife.24001.010
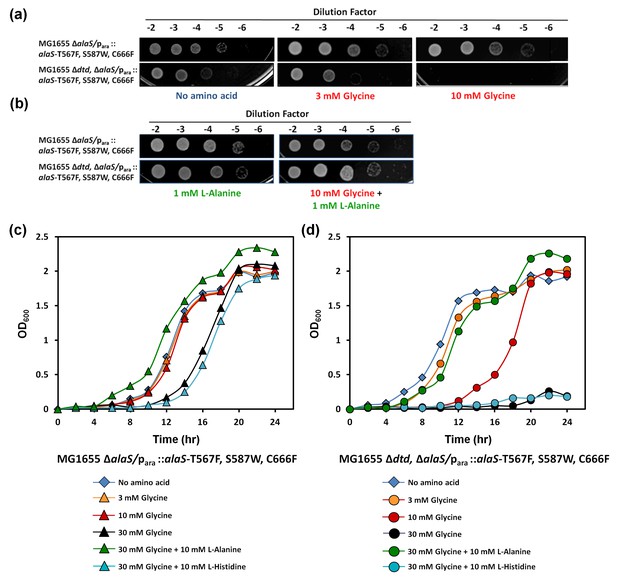
DTD knockout causes pronounced glycine toxicity in E. coli.
Spot dilution assay of E. coli MG1655 ∆alaS/para: : alaS-T567F, S587W, C666F compared with that of E. coli MG1655 ∆dtd, ∆alaS/para:: alaS-T567F, S587W, C666F (a) in the presence of no amino acid, 3 mM glycine, or 10 mM glycine, and (b) in the presence of 1 mM L-alanine, or 10 mM glycine and 1 mM L-alanine. (c) Growth curve of E. coli MG1655 ∆alaS/para: : alaS-T567F, S587W, C666F supplemented with no amino acid (blue diamond), 3 mM glycine (orange triangle), 10 mM glycine (red triangle), 30 mM glycine (black triangle), 30 mM glycine and 10 mM L-alanine (green triangle), 30 mM glycine and 10 mM L-histidine (cyan triangle) (d) Growth curve of E. coli MG1655 ∆dtd, ∆alaS/para: : alaS-T567F, S587W, C666F supplemented with no amino acid (blue diamond), 3 mM glycine (orange circle), 10 mM glycine (red circle), 30 mM glycine (black circle), 30 mM glycine and 10 mM L-alanine (green circle), 30 mM glycine and 10 mM L-histidine (cyan circle).
-
Figure 5—source data 1
Growth curves of E. coli MG1655 with and without dtd knockout in AlaRS editing-defective background.
- https://doi.org/10.7554/eLife.24001.012
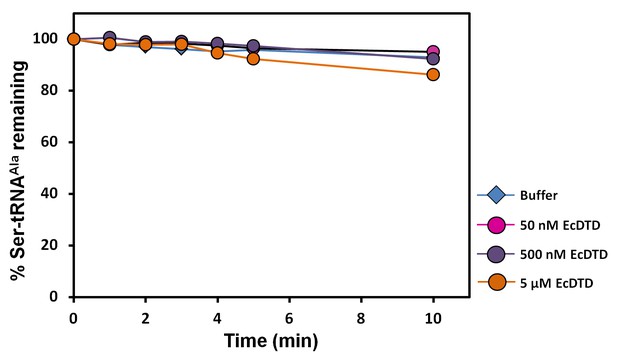
DTD is inactive on Ser-tRNAAla.
Deacylation of Ser-tRNAAla by buffer (blue diamond), 50 nM EcDTD (pink circle), 500 nM EcDTD (purple circle), 5 µM EcDTD (orange circle).
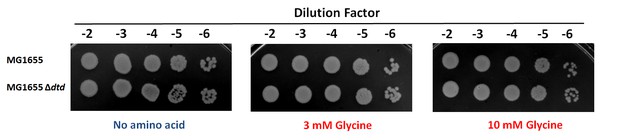
Spot dilution assay of E. coli MG1655 compared with E. coli MG1655 ∆dtd with increasing concentration of glycine.
https://doi.org/10.7554/eLife.24001.014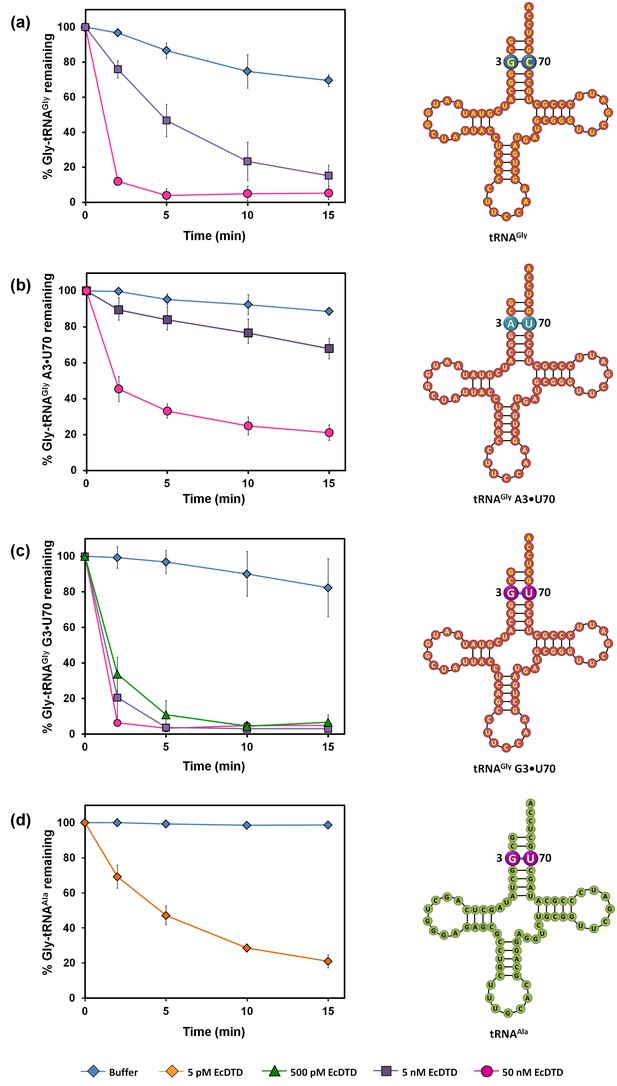
DTD positively selects the tRNA acceptor stem element G3•U70.
(a) Deacylation of Gly-tRNAGly by buffer (blue diamond), 5 nM EcDTD (purple square), 50 nM EcDTD (pink circle). (b) Deacylation of Gly-tRNAGly A3•U70 by buffer (blue diamond), 5 nM EcDTD (purple square), 50 nM EcDTD (pink circle). (c) Deacylation of Gly-tRNAGly G3•U70 by buffer (blue diamond), 500 pM EcDTD (green triangle), 5 nM EcDTD (purple square), 50 nM EcDTD (pink circle). (d) Deacylation of Gly-tRNAAla by buffer (blue diamond), 5 pM EcDTD (orange diamond). Error bars indicate one standard deviation from the mean of triplicate readings.
-
Figure 6—source data 1
Deacylation of Gly-tRNAGly mutants and Gly-tRNAAla by DTD.
- https://doi.org/10.7554/eLife.24001.016
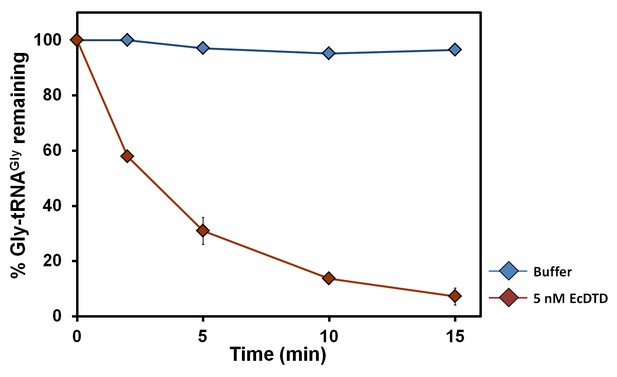
DTD’s activity on the cognate achiral substrate.
Deacylation of Gly-tRNAGly by buffer (blue diamond), 5 nM EcDTD (brown diamond). Error bars indicate one standard deviation from the mean of triplicate readings.
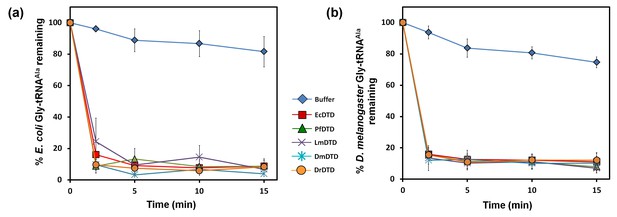
DTD edits Gly-tRNAAla across bacteria and eukaryotes.
(a) Deacylation of E. coli Gly-tRNAAla by buffer (blue diamond), 10 pM EcDTD (red square), 10 pM PfDTD (green triangle), 10 pM LmDTD (purple cross), 10 pM DmDTD (cyan star), 10 pM DrDTD (orange circle). (b) Deacylation of D. melanogaster Gly-tRNAAla by buffer (blue diamond), 10 pM EcDTD (red square), 10 pM PfDTD (green triangle), 10 pM LmDTD (purple cross), 10 pM DmDTD (cyan star), 10 pM DrDTD (orange circle). Error bars indicate one standard deviation from the mean of triplicate readings.
-
Figure 7—source data 1
Deacylation of E. coli Gly-tRNAAla and D. melanogaster Gly-tRNAAla by bacterial and eukaryotic DTDs.
- https://doi.org/10.7554/eLife.24001.019
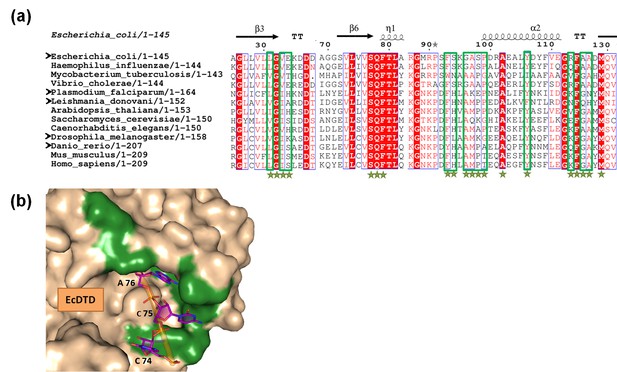
Variations in the tRNA-binding site of DTD.
(a) Structure-based multiple sequence alignment of DTD from different organisms. The residues which are within a distance of 6 Å from the 3′-terminal CCA-arm of tRNA are marked by green stars, and among these, the residues showing variations are enclosed in green box. Black arrowheads indicate the organisms from which DTDs have been tested biochemically in the current study. (b) Surface model of E. coli DTD (PDB id: 1JKE) with 3′-terminal CCA-arm of tRNA placed in the active site on the basis of D-Tyr3AA from the structure of P. falciparum DTD (PDB id: 4NBI). The residues colored in green show the positions showing variations among different organisms.
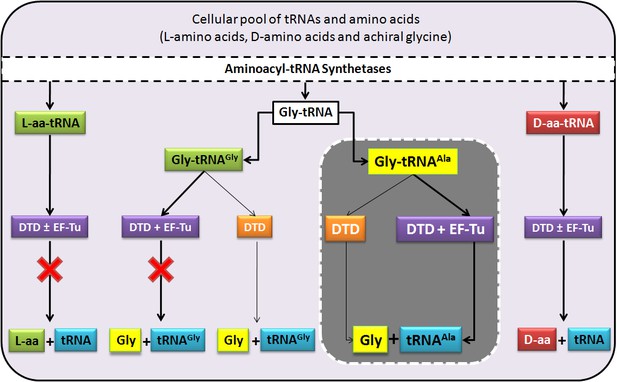
DTD doubles as a key factor to uncouple glycine mischarged on tRNAAla.
In the cell, aminoacylation by aaRSs leads to the formation of different aa-tRNAs, of which L-aa-tRNAs (left extreme) are not acted upon by DTD, while D-aa-tRNAs (right extreme) are effectively decoupled in the presence or absence of EF-Tu, thereby enforcing homochirality. Glycylated tRNAs are acted upon by DTD (centre) but EF-Tu offers protection to the cognate Gly-tRNAGly to prevent its misediting, while the mischarged/non-cognate Gly-tRNAAla is efficiently cleared even in the presence of EF-Tu. Thick connecting arrows indicate the cellular scenario, wherein both DTD and EF-Tu are present.
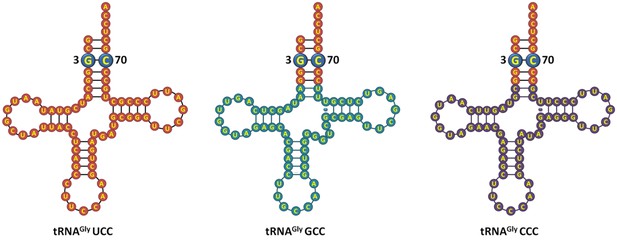
E. coli tRNAGly isoacceptors showing identical acceptor stem.
https://doi.org/10.7554/eLife.24001.023Tables
Protein concentration estimation.
| Protein Name | Concentration of protein taken (mg/ml) measured using A280 | Protein concentration estimation (mg/ml) using | |
|---|---|---|---|
| Bicinchoninic acid Assay | Bradford Assay | ||
| E. coli AlaRS | 4 | 3.82 | 5.22 |
| 8 | 7.45 | 9.02 | |
| E. coli DTD | 4 | 4.36 | 4.83 |
| 8 | 8.08 | 7.93 | |





