Malaria parasite LIMP protein regulates sporozoite gliding motility and infectivity in mosquito and mammalian hosts
Figures
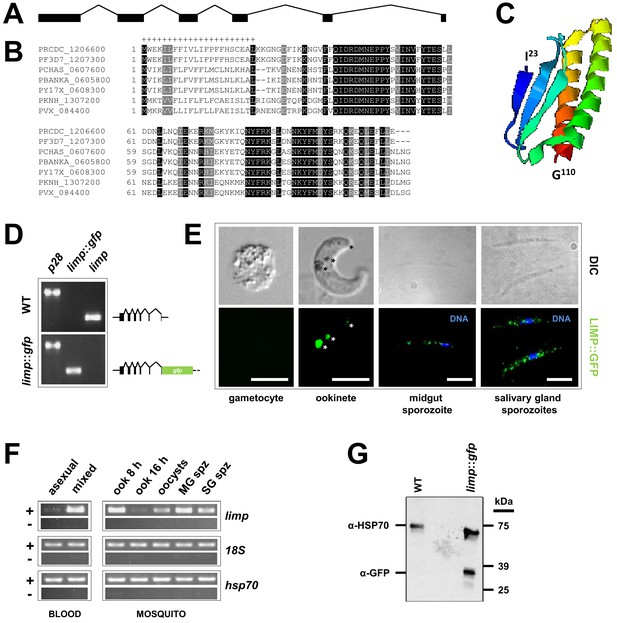
Gene and protein structure of limp and its translational regulation during transmission.
(A) The Plasmodium berghei limp gene (1194 bp) is composed of six exons (shaded bars) and five introns (lines). (B) Protein alignment of LIMP orthologues from seven Plasmodium species. ‘+' signal peptide in P. berghei. (C) QUARK ab initio folding simulation model of LIMP. (D) RT-PCR analyses shows absence of WT limp and presence of correctly spliced limp::gfp mRNA in blood stages of limp::gfp parasites. p28 serves as control transcript. (E) Immunofluorescence assay (IFA) of limp::gfp blood stages shows no LIMP::GFP expression in gametocytes. Live imaging of blood meal-retrieved ookinetes shows LIMP::GFP localisation in discrete foci (*). IFA of limp::gfp midgut sporozoites at day 24 p.i. and salivary gland sporozoites from day 21 p.i. shows a speckled distribution of LIMP::GFP. Parasites were stained with anti-GFP antibodies and DNA was stained with Hoechst-33342. Scale bars = 5 µm. (F) RT-PCR of limp through the parasite life cycle. Asexual: asexual blood stages; mixed: asexuals and gametocytes; ook: ookinetes (8 and 16 hr after gametocyte activation); MG spz: midgut sporozoites; SG spz: salivary gland sporozoites. 18S rRNA and hsp70 serve as loading control genes. +: RT-positive reaction; −: RT-negative reaction. (G) Western Blot analysis of parasites from 20 hr in vitro ookinete culture to verify correct C-terminal GFP tagging of LIMP. Antibodies against GFP (α-GFP) were used to visualise LIMP::GFP (37 kDa) and antibodies against parasite HSP70 (α-HSP70) served as loading control.
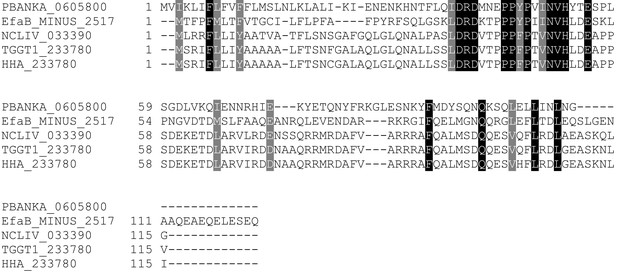
Multiple sequence alignment of LIMP orthologues from related apicomplexan parasites.
EfaB: Eimeria falciformis; NCLIV: Neospora caninum; TGGT1: Toxoplasma gondii; HHA: Hammondia hammondi. All sequences from eupathdb.org.
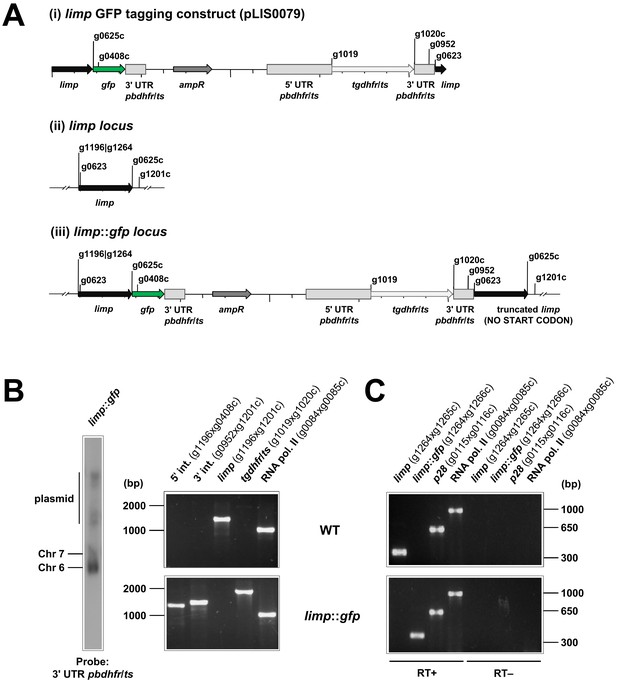
Generation and genotyping of the limp::gfp parasite line.
(A) limp GFP tagging construct pLIS0079 (i) was obtained by cloning the last 1149 bp of the limp ORF excluding the stop codon upstream and in frame with the gfp gene. This construct includes Toxoplasma gondii dhfr/ts selectable marker cassette under the control of P. berghei dhfr/ts 5' and 3' UTRs. The construct was integrated into the limp locus (ii) of cl15cy1 by single homologous recombination, resulting in the fusion of limp to gfp in limp::gfp parasites (iii). (B) Correct tagging of limp was shown by Southern analysis of separated chromosomes (left) and diagnostic PCR analyses (right). Hybridisation of separated chromosomes from uncloned limp::gfp parasites with a probe against the 3' UTR of pbdhfr/ts recognised integrated pLIS0079 into chromosome 6, the endogenous pbdhfr/ts locus in chromosome 7 and circular plasmid with higher molecular weight. PCR analyses in clonal limp::gfp parasites confirm 5' and 3' integration (int.) of pLIS0079, absence of WT limp ORF and presence of tgdhfr/ts gene. (C) Absence of WT limp and presence of limp::gfp mRNA was confirmed in cloned limp::gfp mixed blood stages by RT-PCR. p28 and RNA polymerase II serve as control genes. Primer g1265c only binds to limp WT cDNA while primer g1266c only anneals with limp::gfp cDNA.
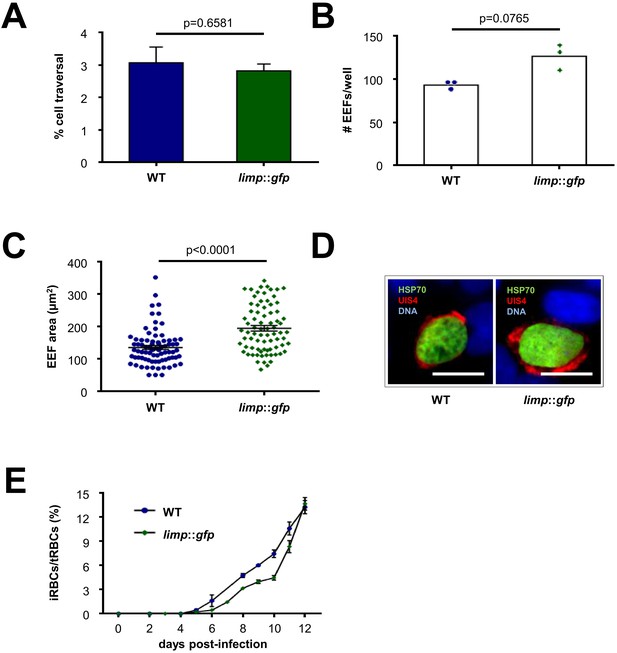
limp::gfp parasites show no defects in liver- and blood-stage infections.
(A) Dextran assay of sporozoite hepatocyte traversal. Bars show means±SEM; p-value for Student´s t-test. WT and limp::gfp (one experiment; n = 3). (B) Exoerythrocytic form (EEF) numbers during in vitro infection of hepatocytes. Bars show means; dots show individual data points. WT and limp::gfp (one experiment; n = 3). (C) Area of exoerythrocytic forms (EEFs) at 45 h p.i. Lines show means±SEM. WT (one experiment; n = 79); limp::gfp (one experiment; n = 77). (B–C) p-values for Mann-Whitney test. (D) Representative images of WT and limp::gfp exoerythrocytic forms at 45 h p.i. Scale bars = 20 µm. (E) Blood-stage infection following mosquito bite. Lines show means ± SEM. WT (two independent experiments; n = 2); limp::gfp (three independent experiments; n = 3). iRBCs: infected red blood cells; tRBCs: total red blood cells.
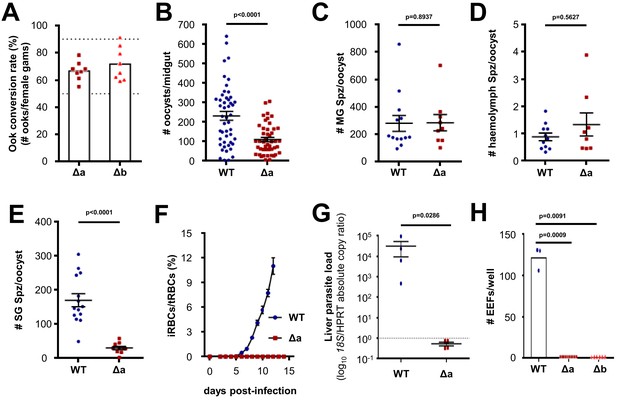
Δlimp parasites suffer cumulative population loss during mosquito and rodent passage.
(A) Ookinete (Ook) conversion rates of both Δa and Δb parasites (50–90% dashed lines). Bars show means, dots represent individual conversion rates. Δa and Δb (four independent experiments, n = 8). (B) Oocysts per mosquito midgut on day 12 p.i. WT (four independent experiments; n = 48); Δa (six independent experiments; n = 46). (C) Midgut sporozoites (MG Spz) per oocyst. WT (three independent experiments; n = 13); Δa (five independent experiments; n = 9). (D) Haemolymph sporozoites (Spz) per oocyst. WT (three independent experiments; n = 11); Δa (four independent experiments; n = 8). (E) Salivary gland sporozoites (SG Spz) per oocyst. WT (four independent experiments; n = 14); Δa (six independent experiments; n = 10). (B–E) Lines show means±SEM; p-values for Mann-Whitney test. (F) Blood-stage infection following i.v. injection of sporozoites. Lines show means ± SEM. WT and Δa (three independent experiments; n = 4). iRBCs: infected red blood cells; tRBCs: total red blood cells. (G) Parasite liver load as measured by qPCR at 44 h.p.i. Mean log10 18S/HPRT absolute copy ratio (two technical replicates) for each mouse is represented by the dots. Lines show means±SEM; p-value for Mann-Whitney test. Note that the number of 18S copies per copy of HPRT in all Δa-infected mice was estimated as being below 1 (dashed line), meaning that Δa parasites do not establish a successful liver infection. (H) Exoerythrocytic form (EEF) numbers during in vitro infection of hepatocytes. Bars show means; dots show individual data points; p-values for Mann-Whitney test. WT (one experiment; n = 3); Δa (four independent experiments; n = 10); Δb (two independent experiments; n = 6).
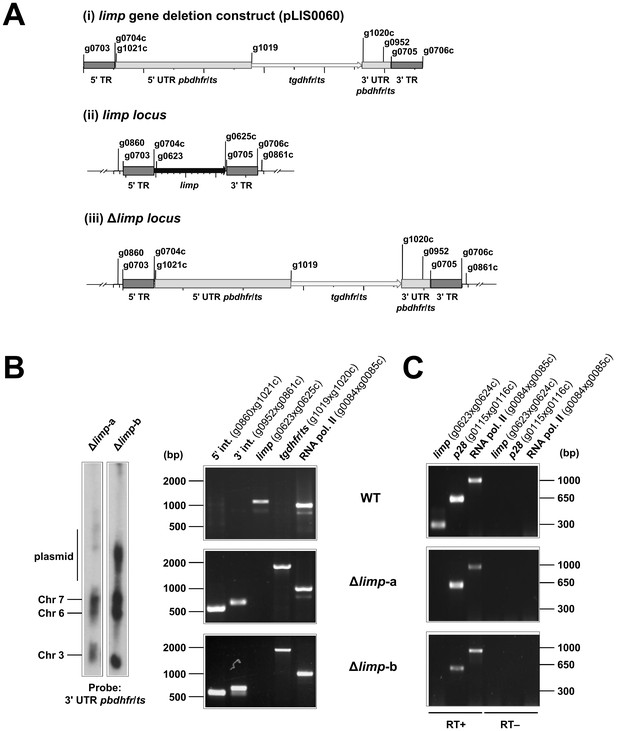
Generation and genotyping of Δlimp parasite lines.
(A) limp gene deletion construct pLIS0060 (i) was obtained by cloning limp 5' and 3' targeting regions (TR) upstream and downstream of the Toxoplasma gondii dhfr/ts selectable marker cassette, respectively. tgdhfr/ts gene is under the control of P. berghei dhfr/ts 5' and 3' UTRs. The construct was integrated into the limp locus (ii) of Fluo-frmg by double homologous recombination, resulting in the complete deletion of limp ORF in Δlimp parasites (iii). (B) Correct deletion of limp was shown by Southern analysis of separated chromosomes (left) and diagnostic PCR analyses (right). Hybridisation of separated chromosomes from uncloned Δlimp parasites with a probe against the 3' UTR of pbdhfr/ts recognised integrated pLIS0060 into chromosome 6, the endogenous pbdhfr/ts locus in chromosome 7, reporter gene constructs (GFP/RFP) in chromosome 3 and circular plasmid with higher molecular weight. PCR analyses in clonal Δlimp parasites confirm 5' and 3' integration (int.) of pLIS0060, absence of limp ORF and presence of tgdhfr/ts gene. (C) Absence of limp mRNA was confirmed in cloned Δlimp mixed blood stages by RT-PCR. p28 and RNA polymerase II serve as control genes. Primer g0624c only anneals with limp cDNA.
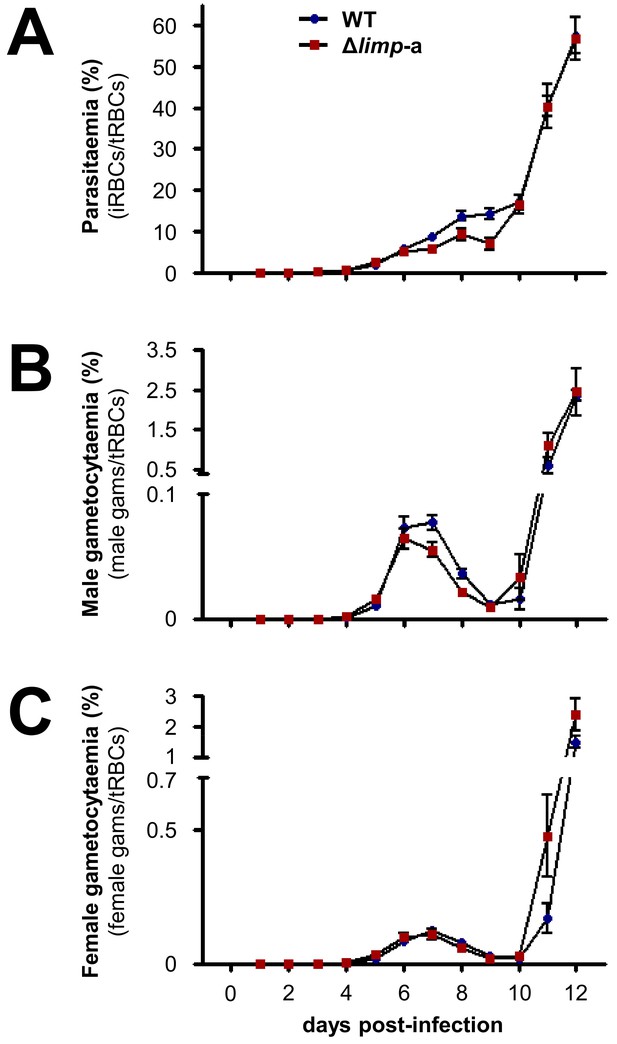
Δlimp parasites develop normal blood-stage parasitaemia and gametocytaemias over the course of infection.
(A) WT and Δlimp-a total parasitaemias over time. (B) WT and Δlimp-a male gametocytaemias over time. (C) WT and Δlimp-a female gametocytaemias over time. (A–C) Lines show means±SEM values from three independent experiments. WT and Δlimp-a (n = 6). iRBCs: infected red blood cells; tRBCs: total red blood cells; gams: gametocytes.
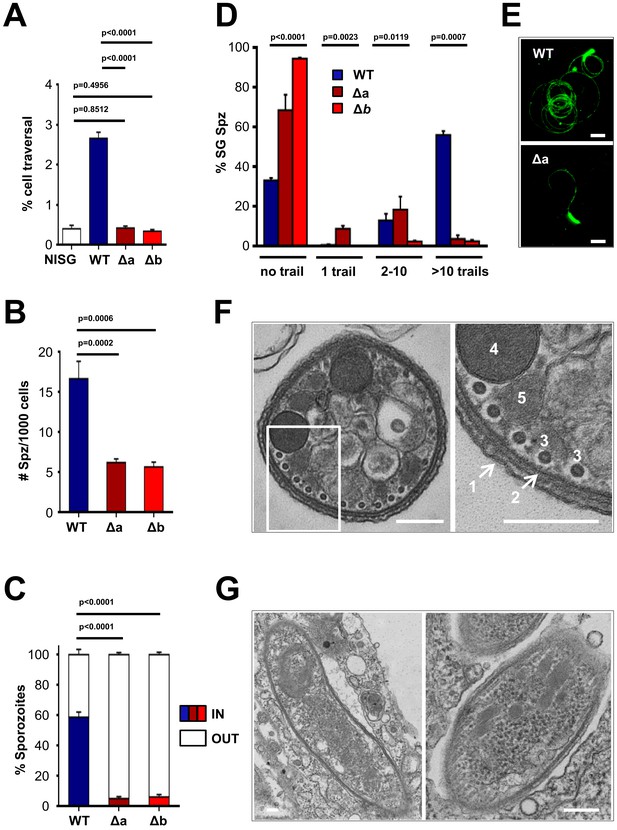
Δlimp parasites are severely impaired in their gliding motility, adhesion and liver cell invasion capabilities.
(A) Dextran assay of sporozoite hepatocyte traversal. NISG: cells incubated with non-infected salivary gland material. WT (four independent experiments; n = 12); Δa (five independent experiments; n = 10); Δb (two independent experiments; n = 6). (B) Sporozoite (Spz) hepatocyte adhesion assay. WT (three independent experiments; n = 6); Δa (three independent experiments; n = 8); Δb (two independent experiments; n = 6). (C) Sporozoite hepatocyte invasion assay. WT (four independent experiments; n = 12; 1292 sporozoites analysed); Δa (five independent experiments; n = 12; 806 sporozoites analysed); Δb (two independent experiments; n = 6; 381 sporozoites analysed). (A–C) Bars show means±SEM; p-values for Student´s t-test. (D) Salivary gland sporozoite (SG Spz) gliding motility assay. Bars show means ± SEM; p-value for Kruskal-Wallis test. WT (four independent experiments; n = 7; 820 sporozoites analysed); Δa (four independent experiments; n = 4; 477 sporozoites analysed); Δb (one experiment; n = 3; 299 sporozoites analysed). (E) Representative images of WT and Δa CSP gliding trails. Scale bars = 10 µm. (F) Cellular ultrastructure of Δa sporozoites is unchanged. (1) plasma membrane; (2) inner membrane complex; (3) subpellicular microtubules; (4) rhoptries; (5) micronemes. (G) Few Δa sporozoites are seen within the mosquito’s salivary gland parenchyma, meaning they actively invaded this tissue and are thus not unviable. (F–G) Scale bars = 200 nm.
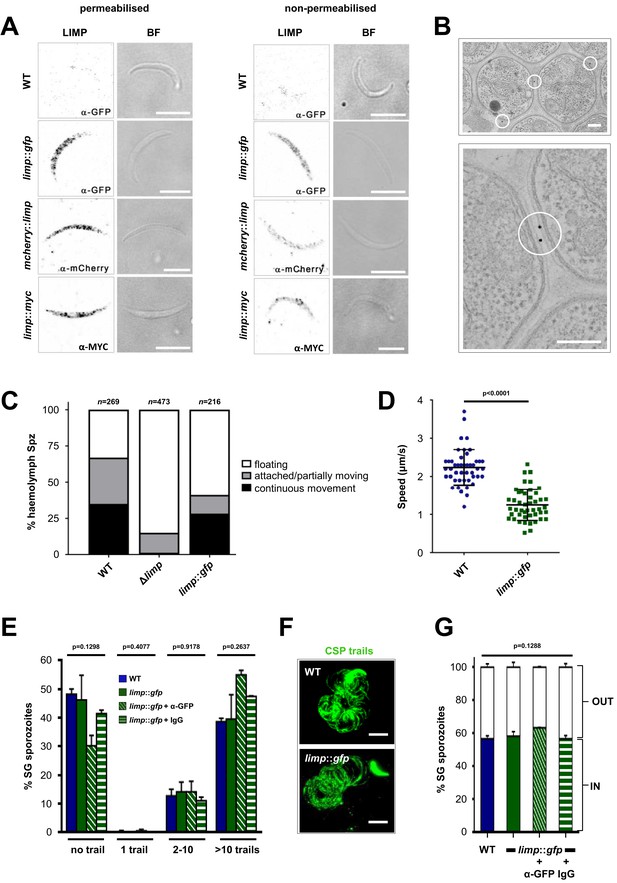
LIMP localises to the parasite plasma membrane but its function cannot be targeted by blocking antibodies.
(A) Immunofluorescence assays of WT, limp::gfp, mcherry::limp and limp::myc salivary gland sporozoites. Sporozoites were stained with anti-GFP (α-GFP), α-mCherry or α-MYC antibodies at days 17–20 p.i. BF=brightfield images. Scale bars = 5 µm. (B) Immuno-electron microscopy of limp::gfp salivary gland sporozoites with anti-GFP antibodies identifies LIMP::GFP (black dots inside white circles) at the parasite plasma membrane. Scale bars = 200 nm. (C) Live motility assay to investigate movement patterns of WT, Δlimp and limp::gfp haemolymph sporozoites (Spz). Most Δlimp sporozoites display a severe attachment phenotype and thus are incapable of moving. The indicated total number of sporozoites derived from three independent experiments in case of WT and six or two independent assays were performed for Δlimp or limp::gfp, respectively. (D) Moving limp::gfp haemolymph sporozoites show reduced gliding speed. Dot plots show means±SEM of 46 analysed sporozoites from three independent experiments per parasite line; p-value for Mann-Whitney test. (E) CSP shedding-based gliding motility assay of WT and limp::gfp salivary gland (SG) sporozoites in the presence and absence of anti-GFP (α-GFP) blocking antibodies or control IgGs. Bars show means±SEM. WT (one experiment; n = 3; 280 sporozoites analysed); limp::gfp (two independent experiments; n = 5; 492 sporozoites analysed); limp::gfp + α-GFP (one experiment; n = 3; 197 sporozoites analysed); limp::gfp + IgG (one experiment; n = 3; 234 sporozoites analysed). (F) Representative images of WT and limp::gfp (no antibody) CSP gliding trails. Scale bars = 10 µm. (G) WT and limp::gfp salivary gland (SG) sporozoite hepatocyte invasion assay in the presence and absence of anti-GFP (α-GFP) blocking antibodies or control IgGs. Bars show means±SEM of invading (IN) and non-invading (OUT) parasites. Data are from one experiment (n = 3) for all parasite lines and conditions. Numbers of sporozoites analysed: WT = 346, limp::gfp = 763, limp::gfp + α-GFP = 807, limp::gfp + IgG = 742. (E and G) p-values for Kruskal-Wallis test.
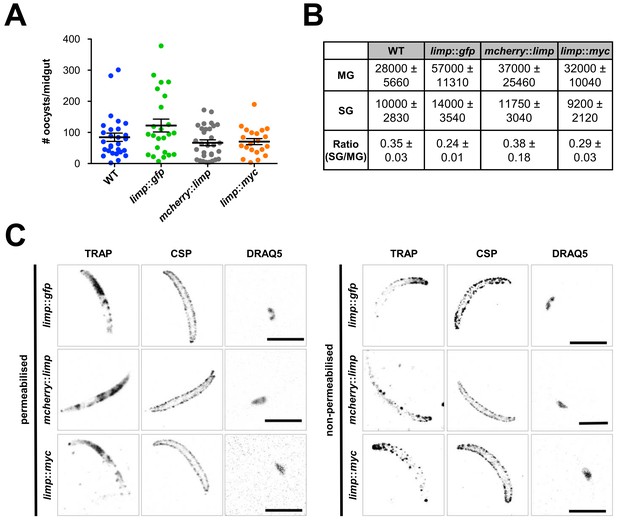
Phenotypic characterisation of tagged limp parasite lines.
Oocysts per mosquito midgut. All tagged lines show oocyst numbers similar to WT parasites indicating no defect in midgut colonisation. Lines show means±SEM from two independent experiments. WT (n = 28); limp::gfp (n = 24); mcherry::limp (n = 31); limp::myc (n = 21). (B) Analysis of salivary gland invasion by determining midgut (MG) and salivary gland (SG) sporozoite numbers on day 17 p.i. Values show means±SD from two independent experiments. (C) Immunofluorescence assays of limp::gfp, mcherry::limp and limp::myc salivary gland sporozoites. Sporozoites were stained for the surface marker CSP and the surface/microneme marker TRAP at days 17–20 p.i. DNA stained with DRAQ5. Scale bars = 5 µm.
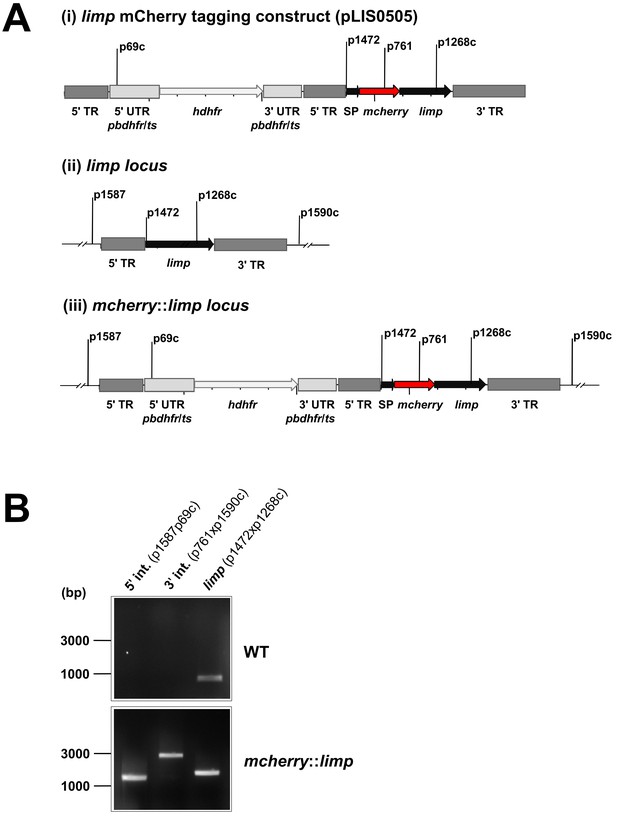
Generation and genotyping of the mcherry::limp parasite line.
(A) mCherry limp tagging construct pLIS0505 (i) was obtained by inserting the mcherry gene downstream and in frame of the signal peptide (SP) of limp. To ensure correct cleavage of the N-terminal signal peptide, the sequence after the cleavage site is integrated again downstream of mcherry followed by the limp ORF and endogenous 3' UTR which, together with the endogenous limp promoter, serve as targeting regions (TRs). This construct includes human dhfr selectable marker cassette under the control of P. berghei dhfr/ts 5' and 3' UTRs. The construct was integrated into the limp locus (ii) of cl15cy1 by double homologous recombination, resulting in replacement of endogenous limp by a N-terminally mCherry-tagged version of limp (iii). (B) Correct tagging of limp was shown by diagnostic PCR analyses. PCR analyses in clonal mcherry::limp parasites confirm 5' and 3' integration (int.) of pLIS0505 and electrophoretic mobility shift of limp ORF due to the presence of the mcherry tag.
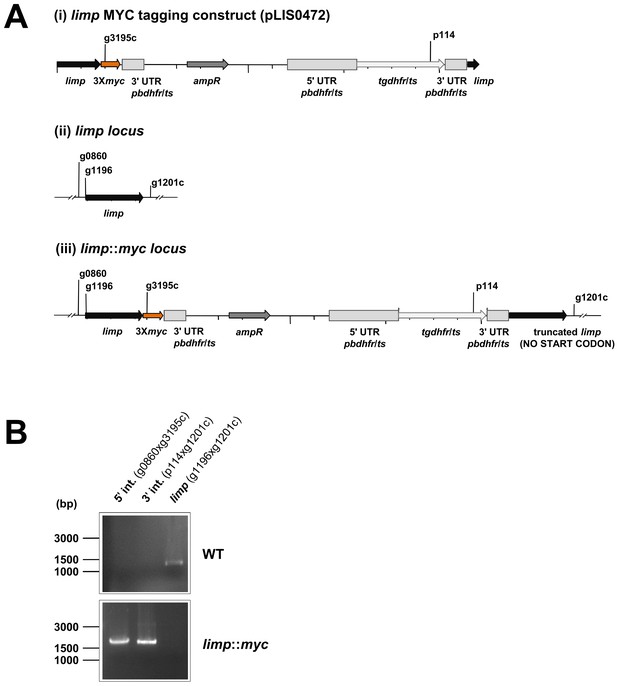
Generation and genotyping of the limp::myc parasite line.
(A) limp MYC tagging construct pLIS0472 (i) was obtained analogously to limp GFP tagging construct by cloning the last 1149 bp of the limp ORF excluding the stop codon upstream and in frame with the triple tandem copy myc tag (3Xmyc). This construct includes Toxoplasma gondii dhfr/ts selectable marker cassette under the control of P. berghei dhfr/ts 5' and 3' UTRs. The construct was integrated into the limp locus (ii) of cl15cy1 by single homologous recombination, resulting in the fusion of limp to myc in limp::myc parasites (iii). (B) Correct tagging of limp was shown by diagnostic PCR analyses. PCR analyses in clonal limp::myc parasites confirm 5' and 3' integration (int.) of pLIS0472 and absence of WT limp ORF.
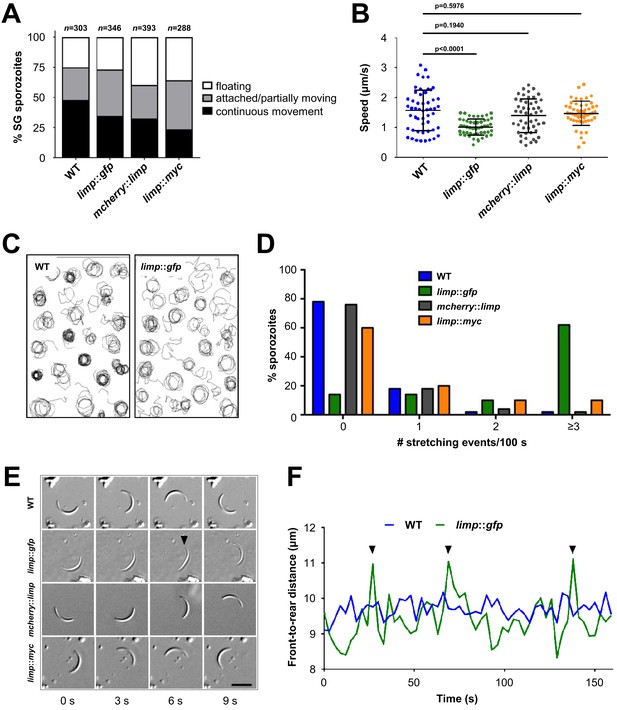
limp::gfp salivary gland sporozoites glide with a ‘limp’.
(A) The percentage of circular moving limp::gfp salivary gland (SG) sporozoites is comparable to WT, mcherry::limp and limp::myc sporozoites. Four independent experiments were performed for each parasite line to obtain indicated numbers of sporozoites. (B) Moving limp::gfp salivary gland sporozoites show significantly reduced gliding speed, whereas neither mcherry::limp nor limp::myc parasites reveal a speed reduction. Lines show means±SD; p-values for one-way ANOVA with Dunnett's post-hoc test. For each line, 50–53 sporozoites were analysed. Data for all lines represent three independent experiments. (C) Maximum projections of a representative subset of WT and limp::gfp moving sporozoites tracked in (B). (D) limp::gfp sporozoites show higher frequency of stretching events compared to WT, mcherry::limp and limp::myc parasites. Percentage of sporozoites within each stretching frequency category are shown by the bars. Fifty sporozoites were analysed per parasite line. Data for all lines are from three independent experiments. (E) Consecutive bright-field images of representative gliding WT, limp::gfp, mcherry::limp and limp::myc sporozoites. Arrowhead indicates the apical tip of a stretched sporozoite. Scale bar = 10 µm. (F) Distance between front and rear ends of one representative sporozoite for WT and limp::gfp, respectively. The distance was analysed over 159 s. Arrowheads indicate stretching events of limp::gfp parasites.
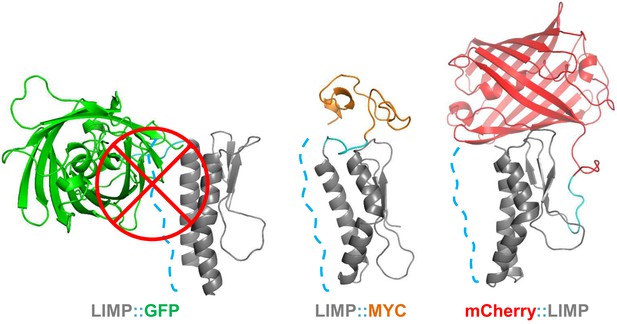
GFP tag occludes solvent accessible surface area of LIMP helices.
Modelled LIMP structures linked to different tags (GFP in green, MYC in orange and mCherry in red) extracted after 100 ns of all-atom molecular dynamics simulations. Solvent accessible surface area of LIMP helices is shown as a blue dashed line. Unlike MYC or mCherry tags that do not interact with LIMP helices, GFP tag interacts with LIMP helices and thus occlude their solvent accessible surface area.
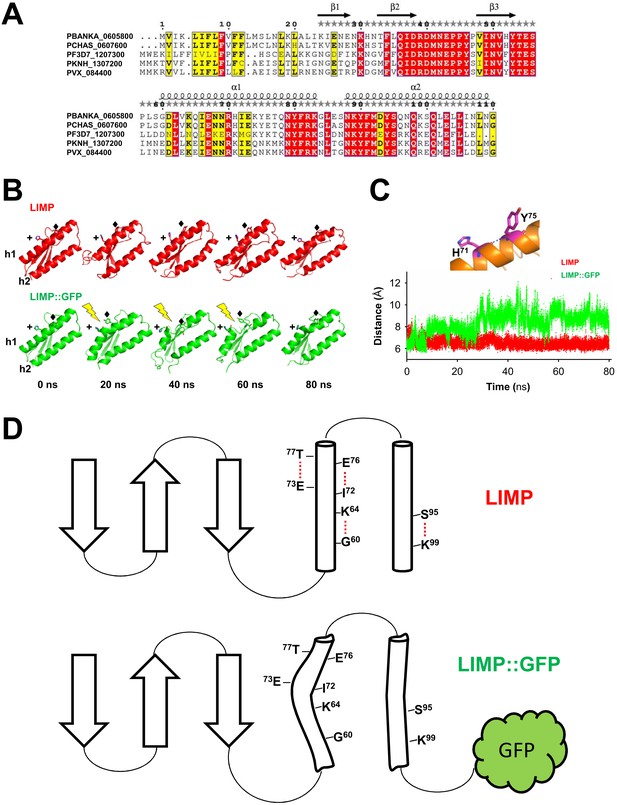
LIMP secondary structure prediction and destabilisation upon GFP tagging.
(A) Multiple sequence alignment of Plasmodium LIMP orthologues with secondary structure prediction shown for P. berghei LIMP. α: alpha helix; β: beta sheet. (B) All-atom molecular dynamics simulations of LIMP and LIMP::GFP at different time points (GFP is not shown for clarity). H71 (+) and Y75 (♦) of helix 1 (h1) are shown as sticks. Loss of helix integrity is indicated in LIMP::GFP by the yellow lightning bolts. (C) Distance plot over time between C-α atoms of H71 and Y75 residues of helix 1 in LIMP and LIMP::GFP simulations. (D) Topology layout of LIMP (top) and LIMP::GFP (bottom) and indication of intrahelical H-bonds that become disturbed in the GFP-tagged fusion protein.
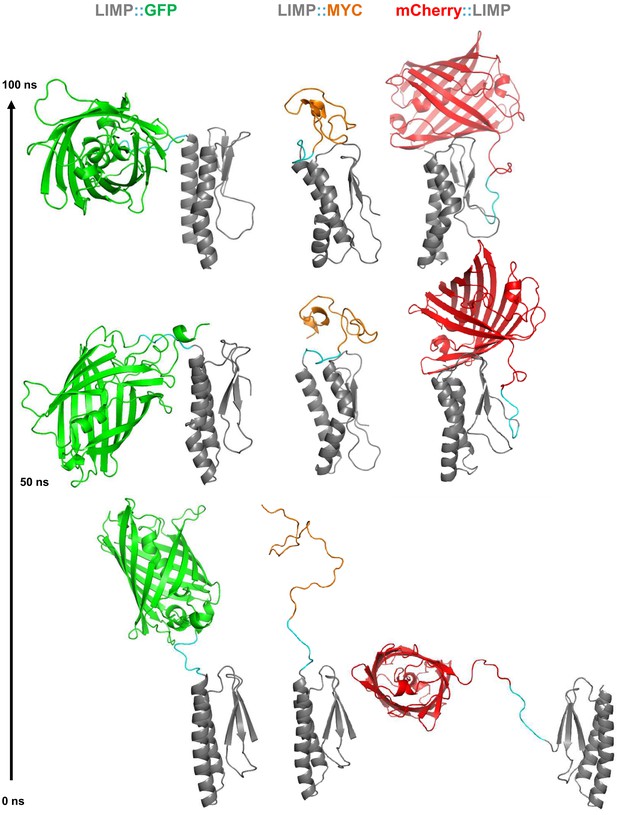
All-atom molecular dynamics simulations of LIMP tagged with GFP, MYC and mCherry.
Starting modelled structures (bottom), stable intermediate structures at 50 ns (middle) and structures after 100 ns (top) are shown. Linker residues between LIMP and tags are highlighted in cyan in all structures.
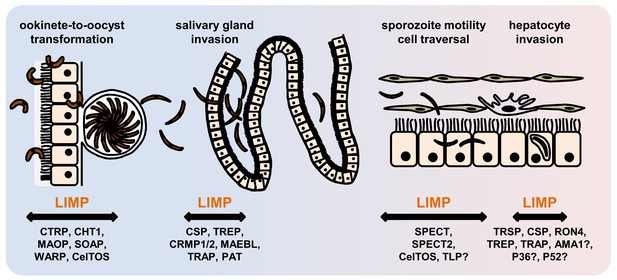
Role of motility- and invasion-related Plasmodium factors during mosquito stage development and transmission of malaria parasites.
While previously published malaria proteins participate in just one or few motility and invasion steps of Plasmodium mosquito stages, LIMP plays a role in midgut invasion/oocyst formation, salivary gland invasion, sporozoite adhesion and gliding, and traversal and invasion of hepatocytes.
Videos
Live gliding motility of a representative wild-type sporozoite
https://doi.org/10.7554/eLife.24109.015Live gliding motility of a representative limp::gfp sporozoite
https://doi.org/10.7554/eLife.24109.016Additional files
-
Supplementary file 1
Summary of Plasmodium surface-, microneme- or rhoptry-localised proteins with function in ookinete and sporozoite motility, adhesion, cell traversal and cell invasion.
- https://doi.org/10.7554/eLife.24109.021
-
Supplementary file 2
Primers used in the present work.
(A) Primers used in life-cycle RT-PCRs and qPCR of RNA extracts from infected livers. (B) Primers used in the generation of gene deletion, GFP-tagging, mCherry-tagging and MYC-tagging constructs. (C) Primers used in genotyping and RT-PCR of mutant parasite lines.
- https://doi.org/10.7554/eLife.24109.022





