O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals
Figures
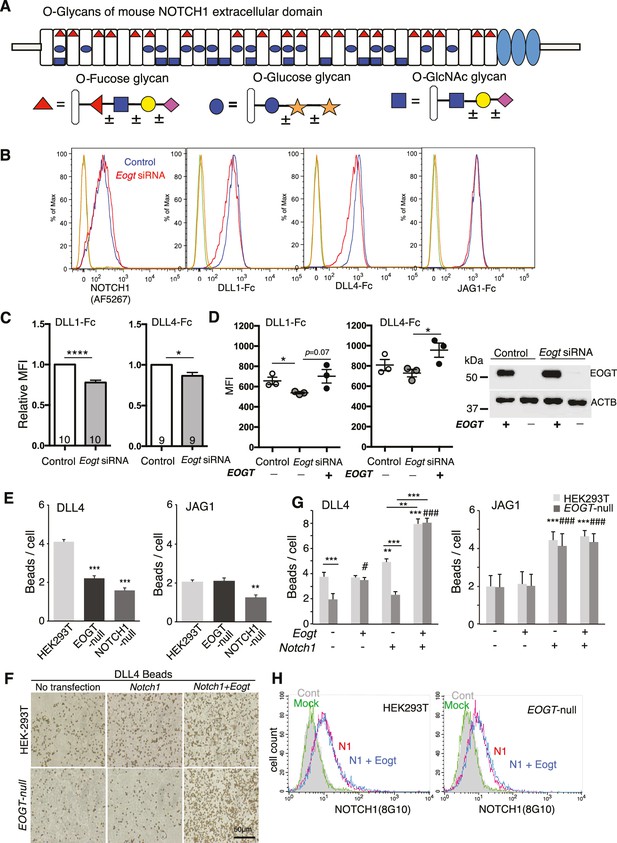
EOGT promotes NOTCH1 binding to Delta ligands.
(A) Diagram of predicted O-glycans on mouse NOTCH1: red triangle, O-fucose glycans; blue circle, O-glucose glycans; blue square, O-GlcNAc glycans. Individual sugar residues that may extend O-fucose, O-glucose or O-GlcNAc to varying degrees are: yellow circle, galactose; pink diamond, sialic acid; orange star, xylose. (B) Flow cytometry of Lec1 CHO cells expressing vector control or Eogt siRNA with NOTCH1 mAb, DLL1-Fc, DLL4-Fc or JAG1-Fc. (C) Relative mean fluorescence index (MFI) for binding of DLL1-Fc and DLL4-Fc to control and Eogt-siRNA Lec1 cells. Concentrations of ligand varied from 100 to 750 ng/ml. MFI values for binding to control cells taken as 1.0 were 455 ± 55 (DLL1-Fc) and 1215 ± 49 (DLL4-Fc). Data from 9 to 10 independent experiments are average, normalized MFI ± SEM; significance determined by paired, two-tailed Student’s t-test, *p<0.05, ****p<0.0001. (D) MFI values obtained for binding of DLL1-Fc or DLL4-Fc (750 ng/ml) to control and Eogt knockdown Lec1 CHO cells, before and after transfection of a human EOGT cDNA. Data are mean ± SEM from three independent experiments. Significance determined by unpaired, two-tailed Student’s t-test, *p<0.05. Western blot analysis of transfectants. (E) DLL4 and JAG1 beads bound to wild-type, EOGT-null, or NOTCH1-null HEK293T cells were observed by microscopy and counted (n = 50). Data are mean ± S.D. from three independent experiments. Statistical analysis was by Welch's t-test. **p<0.01; ***p<0.001. (F) Wild-type or EOGT-null HEK293T cells were transfected with Notch1 alone or together with Eogt followed by incubation with DLL4 beads. The number of DLL4 beads bound to cells was markedly increased by co-transfection of Eogt and Notch1. (G) Wild-type or EOGT-null HEK293T cells or cells transiently transfected with Notch1 with or without Eogt, were incubated with DLL4 or JAG1 beads. The number of Dynabeads bound per transfected cell marked by GFP expression was determined (n = 50). Data are mean ± S.D. from three independent experiments. *p<0.05; **p<0.01; ***p<0.001; #p<0.05; ###p<0.001 compared with the left-most wild type (*) or EOGTnull (#) bar by Welch's t test. (H) Wild-type or EOGT-null HEK293T cells were transfected with Notch1 with or without Eogt and subjected to flow cytometry using 8G10 NOTCH1 Ab. Mock transfectants were analyzed with (Mock) or without primary antibody (Cont).
-
Figure 1—source data 1
Raw data for Figure 1C,D,E,G.
- https://doi.org/10.7554/eLife.24419.004
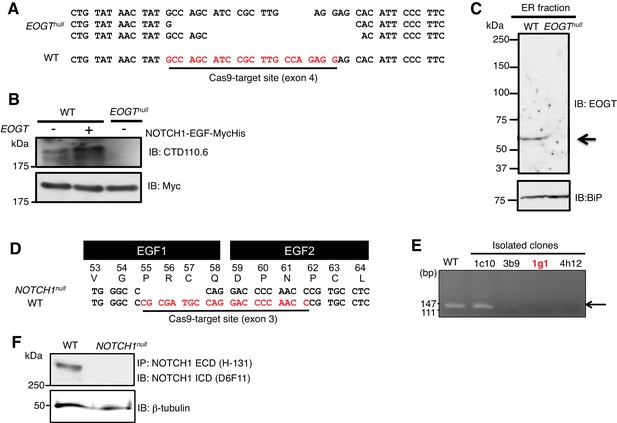
Generation and characterization of EOGT- and NOTCH1-null HEK293T cells.
(A) Schematic diagram of the CRISPR/Cas9 genome editing strategy to generate EOGT-null HEK293T cells. CRISPR/Cas9-mediated DNA cleavage caused frameshift mutations in all three EOGT alleles in HEK293T cells. (B) Wild type or EOGT-null HEK293T cells with Notch1-EGF-mycHis alone or together with EOGT. CTD110.6 immunoblotting revealed the lack of O-GlcNAc on NOTCH1 EGF repeats in the absence of EOGT. (C) Total ER fraction was obtained using Endoplasmic Reticulum Enrichment Extraction Kit (Novus Biologicals [NBP2-29482]) from parental HEK293T cells or EOGT-null cells, and subjected to immunoblotting with EOGT (1:2000 dilution) or BiP (1:10000 dilution) antibodies. (D) Schematic diagram of the CRISPR/Cas9 genome editing strategy to generate NOTCH1-null HEK293T cells. CRISPR/Cas9-mediated DNA cleavage caused frameshift mutations in the single NOTCH1 allele in HEK293T cells. (E) Screening for CRISPR/Cas9-mediated genomic deletion at the Notch1 locus. A clone 1g1 was selected and deletion of the target sequence was confirmed by direct sequencing analysis. (F) Total cell lysates from parental HEK293T cells or NOTCH1-null cells were subjected to immunoprecipitation (IP) using NOTCH1 ECD antibody (H-131). The immunoprecipitates were analyzed by immunoblotting with NOTCH1 ICD antibody (D6F11). Aliquots of total cell lysates were immunoblotted with β-tubulin antibody.
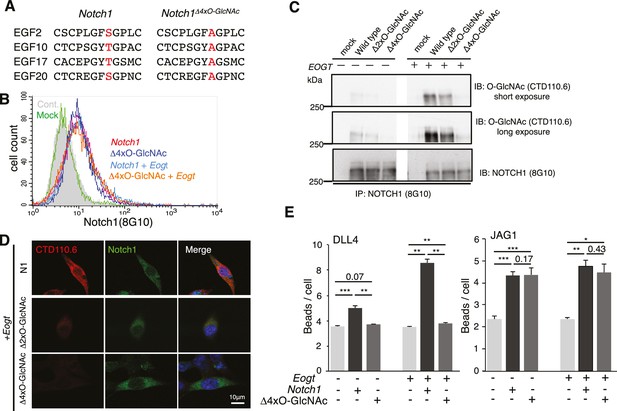
O-GlcNAc on NOTCH1 EGF repeats promotes DLL4-NOTCH1 interactions.
(A) Ala substitution of Thr/Ser in the O-GlcNAc consensus site C5XXG(Y/F)(T/S)GXXC6 in EGF2, 10, 17, and 20 in the NOTCH1Δ4xO-GlcNAc mutant. (B) Cell surface expression of NOTCH1Δ4xO-GlcNAc is comparable to that of wild-type NOTCH1. EOGT-null transfectants expressing Notch1 or Notch1Δ4xO-GlcNAc with or without Eogt were subjected to flow cytometry using 8G10 NOTCH1 antibody. Control transfectants were with (Mock) or without (Cont) primary antibody. Similar results were obtained for wild-type HEK293T cells (not shown). (C) Expression and O-GlcNAcylation of Notch1, Notch1Δ2xO-GlcNAc and Notch1Δ4xO-GlcNAc were analyzed by immunoprecipitation (IP) using NOTCH1 (8G10) antibody, followed by immunoblotting with CTD110.6 O-GlcNAc or NOTCH1 antibodies. (D) HEK293T transfectants expressing NOTCH1, NOTCH1Δ2xO-GlcNAc or NOTCH1Δ4xO-GlcNAc were immunostained for O-GlcNAc (CTD110.6 mAb; red) or NOTCH1 (8G10 mAb; green). Merged images include DAPI (blue) staining. Note that CTD110.6 mAb binding is markedly decreased in the NOTCH1Δ4xO-GlcNAc mutant. (E) Quantification of the number of DLL4- or JAG1-coated Dynabeads bound to Notch1 versus Notch1Δ4xO-GlcNAc transfected cells (marked with GFP), in the presence and absence of Eogt. Data are mean ± S.D from three independent experiments. Each experiment analyzed 50 cells. *p<0.05; **p<0.01; ***p<0.001 (Welch's t test).
-
Figure 2—source data 1
Raw data for Figure 2E.
- https://doi.org/10.7554/eLife.24419.007
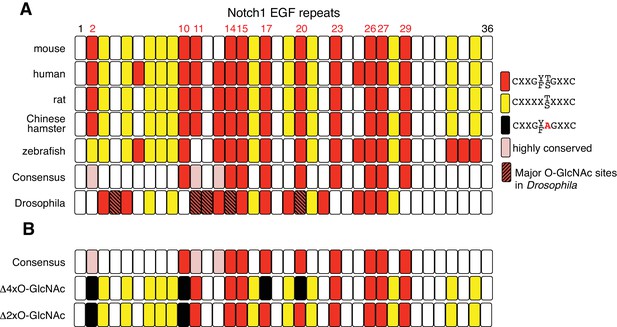
NOTCH1 O-GlcNAc site mutants.
(A) Schematic of the EGF repeats of mammalian NOTCH1 identifying the potential O-GlcNAc consensus site, C5XXG(Y/F)(T/S)GXXC6, on EGF2, 10, 11, 14, 15, 17, 20, 23, 26, 27 and 29. These consensus sites are partly conserved in zebrafish and Drosophila. Recent studies of Drosophila Notch identified EGF4, 11, 12, 14 and 20 as major O-GlcNAcylated sites (Harvey et al., 2016). (B) Schematic of EGF repeats of mouse Notch1Δ2xO-GlcNAc and Notch1Δ4xO-GlcNAc.
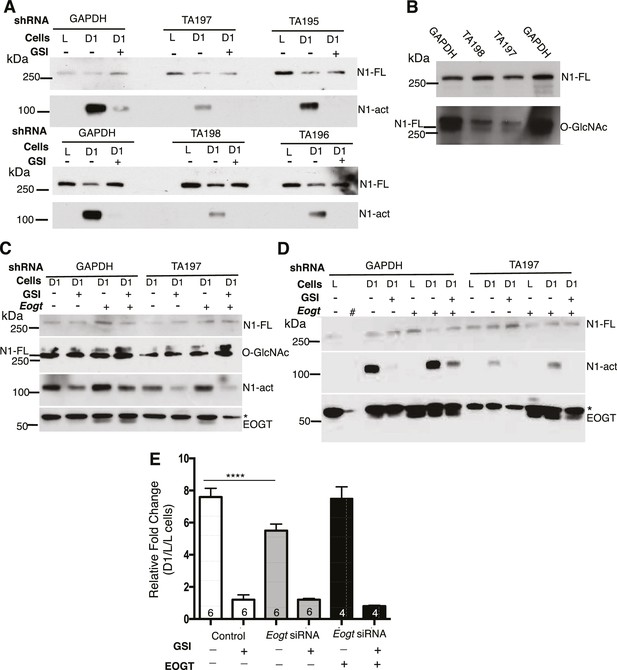
Notch signaling is reduced in EOGT-deficient cells.
(A) Knockdown of EOGT inhibits NOTCH1 activation cleavage. HeLa cells stably expressing shRNAs targeting GAPDH or EOGT (TA195, TA196, TA197, TA198) were co-cultured with L cells or D1/L (D1) cells in the presence and absence of 1 µM DAPT (GSI). After 6 hr, lysates were subjected to Western blot analysis using Abs to detect activated NOTCH1 (N1-act) and NOTCH1 full length (N1–FL) on the relevant section of the PVDF membrane. (B) Western blot analysis of samples from (A) using Ab to detect O-GlcNAc, followed after stripping by Ab to detect N1-FL. (C) HeLa cells stably expressing shRNA against GAPDH or EOGT (TA197) were transfected with vector control or a human EOGT cDNA. After 4 days, co-culture was performed with D1/L (D1) cells in the presence and absence of the DAPT. After ~7 hr, lysates were subjected to Western analysis to detect N1-FL, N1-act and EOGT on the relevant section of the PVDF membrane. O-GlcNAc was detected after stripping the N1-FL membrane section. * non-specific band. (D) The same as (C) except co-culture was with L cells or D1/L cells (D1). The second lane (#) was left empty. * non-specific band. (E) Knockdown of Eogt reduces ligand-induced Notch signaling. Lec1 CHO cells stably expressing siRNAs targeted against Eogt were transfected with TP1-luciferase and TK-renilla luciferase, co-cultured for 30 hr with L cells or D1/L cells, with and without GSI IX (12.5 μM) or a human EOGT cDNA, and dual firefly and renilla luciferase assays were performed. Normalized firefly luciferase activity in L versus D1/L cell co-cultures was plotted as fold-change. The number of independent experiments, each performed in duplicate, are shown in the histogram. Error bars are mean ± standard error and p values were determined by two-tailed paired Student’s t-test (****p<0.0001).
-
Figure 3—source data 1
Raw data for Figure 3E.
- https://doi.org/10.7554/eLife.24419.010
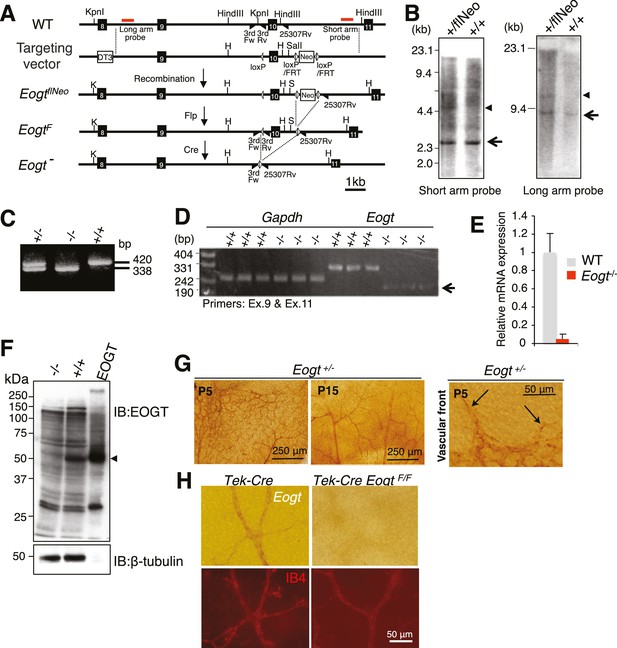
Generation of Eogt-targeted mice.
(A) Schematic drawing of the wild-type mouse Eogt allele (WT), the targeting vector, the floxed allele with the neomycin (Neo)-resistance gene (EogtflNeo), the floxed allele without Neo (EogtF), and the deleted allele (Eogt−). Eogt exons (closed boxes); Neo and diphtheria toxin (DT3) genes served as positive and negative selection markers, respectively (open boxes); loxP sites (gray triangles); FRT sites (open triangles); KpnI (K), HindIII (H), and SalI (S) restriction sites. Homologous recombination between the WT allele and the targeting vector generated the EogtflNeo allele. The EogtF and Eogt− alleles were obtained by Flp-mediated and Ayu1-Cre-mediated recombination, respectively. Red lines indicate positions of probes used for Southern blotting. Positions of primers used for genotyping are indicated by closed triangles. (B) HindIII- or KpnI/SalI-digested genomic DNA isolated from WT (+/+) or heterozygous floxed neo (+/flNeo) ES cells was analyzed by Southern blotting using the short arm or long arm probe, respectively. In addition to signal from the WT allele (arrow), the flNeo mouse shows an additional band corresponding to the recombinant allele (arrowhead). (C) Genomic DNA isolated from WT, Eogt+/−, and Eogt−/− mice was subjected to genotyping using 3rdloxFw, 3rdloxRv, and 25307Rv primers. (D) Semi-quantitative RT-PCR analysis of total RNA from WT or Eogt−/− brain ECs using primers targeting exons 9 and 11. Minor transcripts lacking exon 10 were detected in Eogt−/− mice (arrow). Gapdh was amplified as an internal control. (E) Quantification of Eogt transcripts in Eogt−/− and WT mouse. qRT-PCR analysis of brain ECs showed a marked decrease in Eogt transcripts. Data are mean ± S.D. from three independent experiments performed in triplicate. Each experiment analyzed pooled total RNA obtained from 10 mice. (F) Lack of EOGT protein expression in Eogt−/− mouse. Lung lysates prepared from adult WT or Eogt−/− mice were analyzed in parallel with cell lysates from HEK293T cells overexpressing Eogt. Immunoblotting was performed using anti-EOGT or β-tubulin antibodies. (G) Whole-mount in situ hybridization for Eogt in the P5 or P15 retina. Vascular staining of Eogt is evident in Eogt+/− retina. Eogt expression in vascular sprouts is indicated by arrows. (H) P5 control Tek-Cre and Tek-Cre EogtF/F retinas were subjected to in situ hybridization. Counter staining with Dylight 594-conjugated IB4 is shown below.
-
Figure 4—source data 1
Raw data for Figure 4E.
- https://doi.org/10.7554/eLife.24419.012
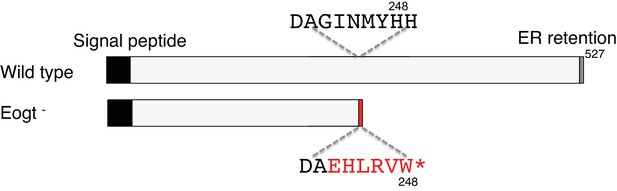
Deletion of exon 10 in the Eogt gene causes exon nine to be spliced to exon 11 leading to a frame shift encoding six novel amino acids before a stop codon.
Truncated EOGT protein was not detected in lung indicating nonsense-mediated decay of mutant transcripts.
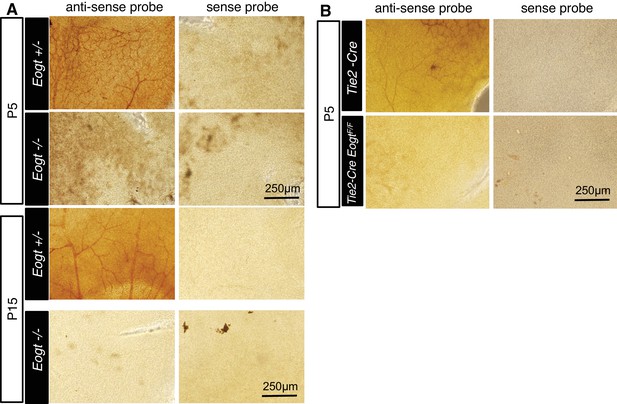
Whole-mount in situ hybridization for Eogt.
(A) In situ hybridization was performed in Eogt+/− or Eogt−/− retinas using anti-sense or sense probes. (B) P5 control Tek-Cre and Tek-Cre EogtF/F retinas were subjected to in situ hybridization using anti-sense or sense probes. Scale bars for (A) and (B), 250 μm.
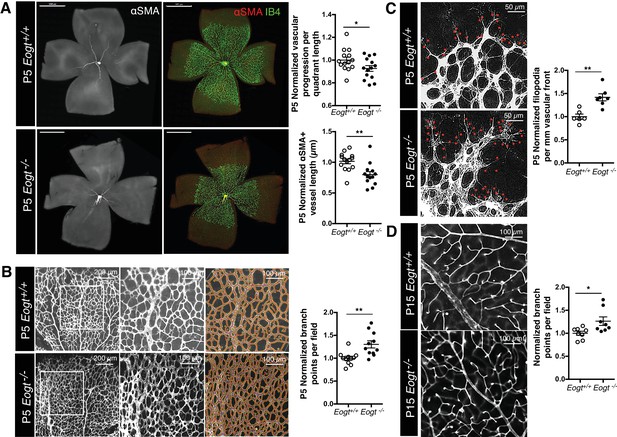
EOGT regulates retinal angiogenesis.
(A) Whole-mount images of Eogt+/+ or Eogt−/− P5 retinas stained with IB4 and anti-αSMA antibody. Bars represent 1000 μm. Scatter plots at right show vascular progression per quadrant length, per retina, and αSMA+ vessel length from the optic nerve per retina, normalized to Eogt+/+ retinas (taken as 1.0). Each symbol represents average vessel progression from 3 to 4 quadrants, or the average length from the optic nerve of 3–4 αSMA+ vessels, per retina. For Eogt+/+, average vascular progression was 0.63 ± 0.03, and the average αSMA+ vessel length was 962 ± 58 µm. (B) Images of P5 vascular front of Eogt+/+ or Eogt−/− retinas stained with IB4. Scatter plot at right shows the normalized number of vascular branch points (500 × 500 µm field (n = 3–8 fields per retina), N = 11 mice). Each symbol represents the average per retina, per mouse. The average Eogt+/+ branch points were 302 ± 27 per retina, taken as 1.0 for normalization. (C) Images of filopodia emanating from tip cells (red dots) at the vascular front. The scatter plot at right shows the number of filopodia in P5 retinas (250 × 250 µm field (n = 4–12 fields per retina), N = 6 mice). The average filopodia per mm vascular front for Eogt+/+ was 33 ± 2 per mm, taken as 1.0 for normalization. (D) Images of P15 retinas stained with IB4. The scatter plot at right shows number of branch points in P15 retinas (500 × 500 µm field (n = 3–8 fields per retina), N = 8 mice). The average for Eogt+/+ was 74 ± 3 per field. Data were normalized from mean ± standard error; p values were calculated by unpaired two-tailed Student’s TTEST. *p≤0.05; **p≤0.01; ***p≤0.001.
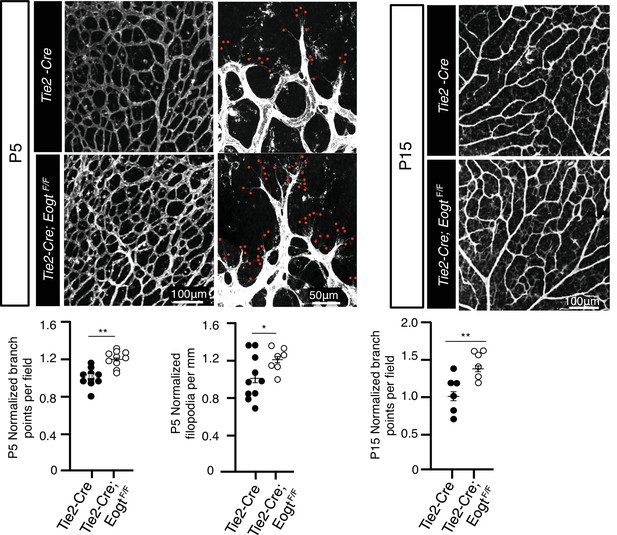
Images of control Tek-Cre and Tek-Cre EogtF/F retinas stained with isolectin B4 (IB4) and quantification of branch points in P5 (N = 6 mice) and P15 (N = 3 mice) retinas and numbers of filopodia per mm vascular front in P5 retinas (N = 6 mice).
Data were normalized from mean ± standard error; p values were calculated by unpaired two-tailed Student’s t-test. *p≤0.05; **p≤0.01.
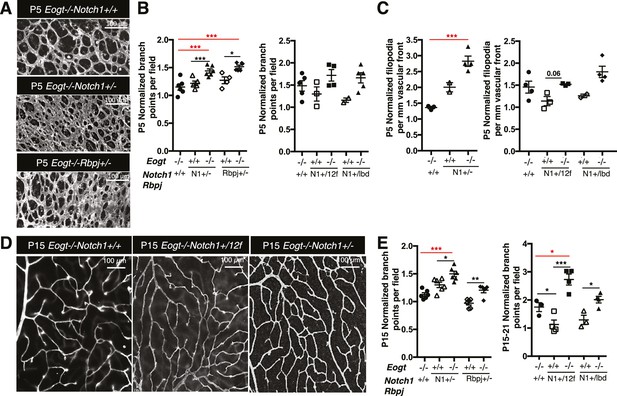
Reduced Notch signaling augments the loss of Eogt in retinal angiogenesis.
(A) Images of vessel density in P5 retinas from Eogt−/− compared to compound mutant mice. (B) Scatter plots represent branch point numbers in P5 retinas from Eogt−/− compared with compound mutant mice, normalized to Eogt+/+ mice. The average number of branch points for the Eogt+/+ used to compare mice with an N1− or Rbpj− allele was 237 ± 5, and for the Eogt+/+ compared to mice with an N12f or N1lbd allele was 380 ± 36 (500 X 500 μm field (n = 3–8 fields per retina), N = 2–6 mice). (C) Scatter plots showing the number of filopodia in P5 compound mutant mice as compared to Eogt−/− mice, normalized to Eogt+/+ mice. Each symbol represents the average number of filopodia per mouse (250 × 250 µm field (n = 4–12 fields per mouse), N = 2–4 mice. The average for Eogt+/+ compared to mice with a N1- or Rbpj- allele was 30 ± 1 per mm and for mice with a N112f or N1lbd allele was 36 ± 4 per mm, taken as 1.0 for normalization. (D) Images of branch points in P15 retinas comparing Eogt−/− to compound mutant mice as indicated. (E) Scatter plots shows branch points in compound mutants compared to Eogt−/− P15 retinas, normalized to Eogt+/+ mice. The average of the Eogt+/+ used to compare mice with a with a N1− or Rbpj− allele was 73 ± 3, and for mice with a N112f or N1lbd allele it was 69 ± 12 (500 x 500 µm field (n = 3–8 fields per retina), N = 3–6 mice. Data represent mean ± standard error except for P5 Eogt+/+Notch1+/− filopodia and P5 Eogt+/+Notch1+/lbd branch points and filopodia that are represented as mean ± range; p values were calculated by unpaired two-tailed Student’s t-test. *p≤0.05; **p≤0.01; ***p≤0.001.
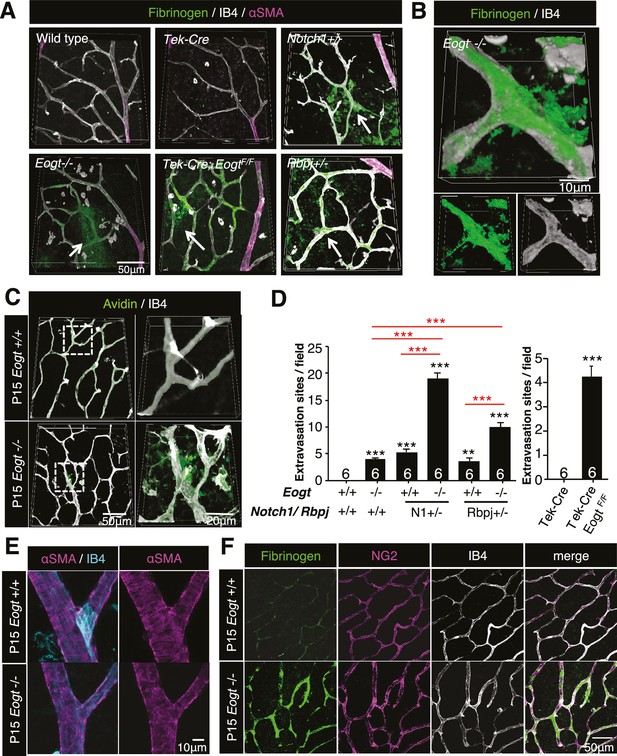
Reduced vessel integrity in the Eogt−/− retina.
(A) Immunostaining with fibrinogen (green) and α-SMA (magenta) antibodies in P15 wild-type, Eogt−/−, Tek-Cre, Tek-Cre:EogtF/F, Notch1+/−, and Rbpj+/− retinas. Arrows indicate fibrinogen staining outside vessels stained by IB4 (white). Three-dimensional images were constructed from confocal images by maximum intensity projection. (B) Higher magnification three-dimensional images of Eogt−/− retina constructed from confocal images using the Alpha-blend method. Below, single channel images showing fibrinogen (green) and IB4 (white) staining. (C) Sulfo-NHS-LC-biotin was perfused into P15 wild-type and Eogt−/− mice and extravasation determined immediately after perfusion by staining with CF488A-conjugated streptavidin (green) and Dylight594-conjugated IB4 (white). Three-dimensional reconstructions were created by maximum intensity projection. Enlarged images of boxed area are shown (right). (D) Sulfo-NHS-LC-biotin was perfused into P15 wild-type, Eogt−/−, Notch1+/−, Eogt−/−Notch1+/− mice as in (C). Quantification of the number of extravasation sites in 210 × 210 μm squares (n = 6 per retina per mouse) is shown. Note that sulfo-NHS-LC-biotin extravasation in Eogt−/− retina is augmented in compound mutant mice. Data represent mean ± standard error; p values determined by Welch's t test. ***p≤0.001. (E) Whole-mount images of wild-type or Eogt−/− P15 retinas stained with IB4 (cyan) and anti-αSMA (magenta) antibody. (F) Whole-mount staining of wild-type and Eogt−/− P15 retinas using IB4 (white) together with anti-fibrinogen (green) and anti-NG2 (magenta) antibodies.
-
Figure 7—source data 1
Raw data for Figure 7D.
- https://doi.org/10.7554/eLife.24419.019
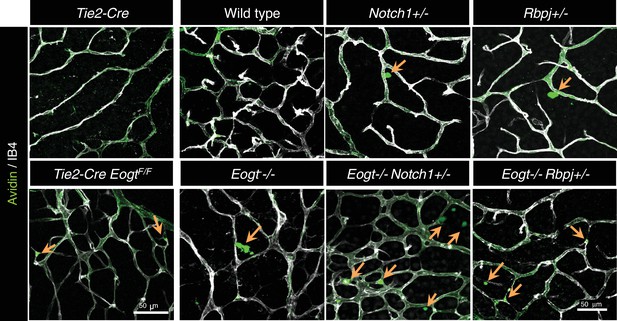
Sulfo-NHS-LC-biotin was perfused into P15 wild-type, Eogt−/−, Tek-Cre, Tek-Cre;EogtF/F, Notch1+/−, Rbpj+/−, Eogt−/−Notch1+/−, Eogt−/−Rbpj+/− mice and stained with CF488A-conjugated streptavidin (green) and Dylight594-conjugated IB4 (white) immediately after perfusion.
The extravasation sites are shown by arrows.
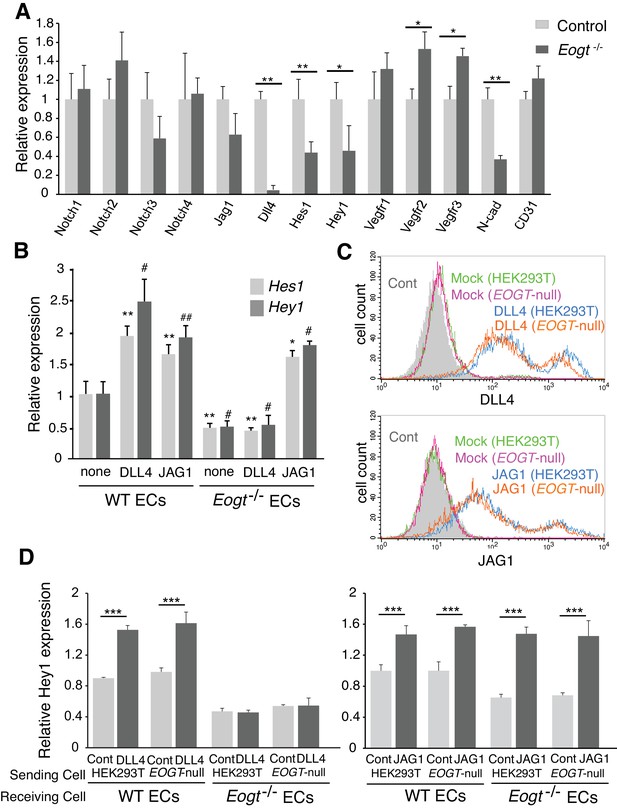
EOGT acts on Notch receptors to regulate ligand-induced Notch signaling in ECs.
(A) Relative mRNA expression in purified brain EC cells. WT (gray) and Eogt−/− (dark gray) EC cells isolated from cerebrum using anti-CD31 antibody were analyzed for gene expression related to the Notch signaling pathway. Gene transcript levels were normalized to Gapdh. Data are mean ± S.D. from three independent experiments performed in triplicate. Each experiment analyzed pooled total RNA obtained from 10 mice. *p<0.05; **p<0.01 (Welch's t test). (B) qRT-PCR analysis of Hes1 and Hey1 in wild type or Eogt−/− lung EC cells following stimulation with immobilized DLL4-Fc or JAG1-Fc. Gene transcripts were normalized to Gapdh. Data are mean ± S.D. from three independent experiments performed in triplicate. Each experiment analyzed total RNA obtained from a single mouse. Significance determined by Welch's t-test. *p<0.05; **p<0.01; #p<0.05; ##p<0.01 compared with the left-most Hes1 (*) or Hey1 (#) histogram. (C) Cells were transfected to express Dll4 or Jag1 and subjected to flow cytometry using DLL4 or JAG1 antibody, respectively. Comparison with mock-transfected cells indicated the amount of exogenously expressed DLL4 and JAG1 on the cell surface in wild-type or EOGT-null HEK293T cells. Mock-transfected HEK293T cells were labeled without primary antibody (Cont.). (D) Dll4/Jag1-transfected cells or Mock transfectants (signal-sending cells) were co-cultured with wild type or Eogt−/− lung EC cells (signal-receiving cells). qRT-PCR analysis of mouse Hey1 expression suggested that EOGT is required for the ability to receive DLL4-mediated Notch signaling, and that EOGT is dispensable for DLL4 and JAG1 as inducers of Notch signaling. Gene transcript levels were normalized to PECAM1 (CD31). Data are mean ± S.D. from three independent experiments performed in triplicate. Significance determined by Welch's t-test. ***p<0.001.
-
Figure 8—source data 1
Raw data for Figure 8A,B,D.
- https://doi.org/10.7554/eLife.24419.022
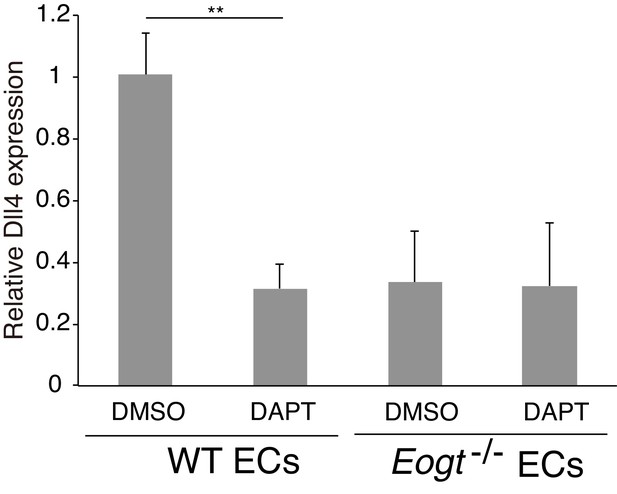
Dll4 expression is suppressed by inhibiting Notch signaling in ECs.
Cultured lung ECs from wild-type or Eogt−/− lungs were treated with DAPT (2 μM) or DMSO for 16 hr. Dll4 expression was analyzed by qRT-PCR. Gene transcripts were normalized to Gapdh. Data are mean ± S.D. from three independent experiments performed in triplicate. Each experiment analyzed total RNA obtained from a single mouse. Significance was calculated using Welch's t test. **p<0.01.
Tables
Primers used for cloning DNA fragments, screening ES cells, or genotyping by PCR.
| Target region Exon10 Fw 5’–ATACGAAGTTATCACCGAACCTAGCCCATATTT–3’ Exon10 Rv2 5’–ACGAAGTTATGTCGACGACTGAGCATTGCTGTT–3’ |
| Long arm region Long arm Fw1 5’–CGAATCAAGCTGTTTGGTCCATTCTCTGCTCCA–3’ Long arm Rv 5’–ACGAAGTTATGGTACGGTCAACTTGAAGAAGTA–3’ |
| Short arm region Short arm Fw 5’–TAGGAACTTCCTCGAAATTCAGTGCTTAGAAGT–3’ Short arm Rv1 5’–GCGCGCCTTTCTCGAACACTGTGTACAGTGACA–3’ |
| Long-arm probe larm16380Fw 5’–CTGCCTCAGCTTCCTGAGTG–3’ larm17196Rv 5’–CATGTCAGATCAGACAGTTC–3’ |
| Short-arm probe sarm26294Fw 5’–CTGAGCTATGTACTGGATGC–3’ AscI-sarmRv2 5’–TGAAGAGGCGCGCCCAGAGACAGAAAAAGCAC–3’ |
| ES cells first screening PGK S1 5’–CCTCCCCTACCCGGTAGAATTGACC–3’ GL1 typing RV2 5’–GAACTGTCAGATTTGGTGACACAGAAAGGC–3’ |
| ES cells second screening 3rdlox Fw 5’–CCACCCGACCCCTGCCAGAACATAATGCTCTCTTGCATC–3’ 3rdlox Rv 5’–GCTGTCGCCAGAGGAGAGAGTGGGTGCTTACTTAC–3’ |
| Eogt mice genotyping 3rdloxFw 5’–CCACCCGACCCCTGCCAGAACATAATGCTCTCTTGCATC–3’ 3rdloxRv2 5’–GCTGTCGCCAGAGGAGAGAGTGGGTGCTTACTTAC–3’ 25307Rv 5’–CCAAGGCGGTCTTGGCCCAT–3’ |
| RT-PCR for Eogt Exon 9 Fw 5’– AGGCTACACGCAGCTCAATT –3’ Exon 11 Rv 5’– AGAAGCCGTGTTTTCGTTGC –3’ |
| qRT-PCR for Eogt Exon 1 Fw 5’–AAGCTGCAGGTCCGTGAAAA–3’ Exon 2 Rv 5’–TAGGTTAGGCTACCGCGTCT–3’ |
-
Underlined sequences are 15 bp homologous overlaps required for In-Fusion cloning.
Primers used for qRT-PCR.
| Target | Forward | Reverse |
|---|---|---|
| Notch1 | AGTGTGACCCAGACCTTGTGA | AGTGGCTGGAAAGGGACTTG |
| Notch2 | CCCAAGGACTGAGAGTCAGG | GGCAGCGGCAGGAATAGTGA |
| Notch3 | ATTTGAGGGGTGCTGAAGTG | GAAGGCTGGGACAGAGAGAA |
| Notch4 | TCCGGACTTTTAAGGCCAAA | TTCCCATTGCTGTGCATACTCT |
| Dll4 | CCCACAATGGCTGTCGTCAT | AACCCTTTGGCCCACTGTTG |
| N-cadherin | AGGGTGGACGTCATTGTAGC | CTGTTGGGGTCTGTCAGGAT |
| CD31 | AGCCAACAGCCATTACGGTTA | AGCCTTCCGTTCTCTTGGTG |
| GAPDH | GGTGCTGAGTATGTCGTGGA | CCTTCCACCATGCCAAAGTT |
| Jag1 | TCTCTGACCCCTGCCATAAC | TTGAATCCATTCACCAGATCC |
| Hes1 | ACACCGGACAAACCAAAGAC | CGCCTCTTCTCCATGATAGG |
| Hey1 | CATGAAGAGAGCTCACCCAGA | CGCCGAACTCAAGTTTCC |
| Hey1 (mouse specific) | TGAATCCAGATGACCAGCTACTGT | TACTTTCAGACTCCGATCGCTTAC |
| Vegfr1 | ACATTGGTGGTGGCTGACTCTC | CCTCTCCTTCGGCTGGCATC |
| Vegfr2 | GCGGGCTCCTGACTACAC | CCAAATGCTCCACCAACTCTG |
| Vegfr3 | CCGCAAGTGCATTCACAGAG | TCGGACATAGTCGGGGTCTT |





