Structure and topology around the cleavage site regulate post-translational cleavage of the HIV-1 gp160 signal peptide
Figures
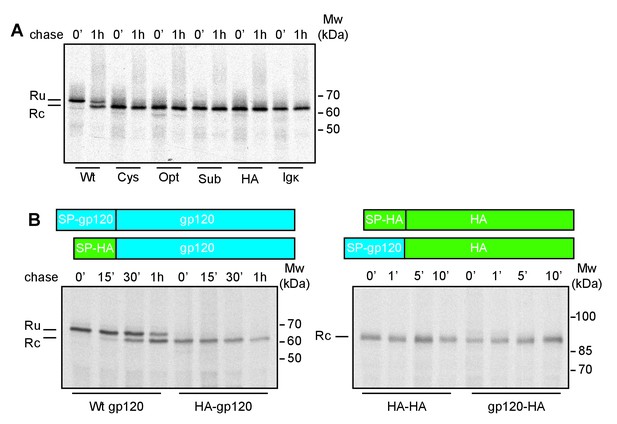
The interplay of gp120 and its natural signal peptide causes post-translational cleavage.
(A) HeLa cells expressing wild-type gp120 (Wt) and gp120 with exogenous signal peptides, HA (HA), Ig κ (Ig κ), cystatin (Cys), Optimal (Opt), and Suboptimal (Sub), were radiolabeled for 10 min and chased for 1 hr or not (0'). After immunoprecipitation gp120 samples were deglycosylated and analyzed using reducing 7.5% SDS-PAGE. Gels were dried and exposed to Kodak-MR films. (B) As in (A) except Wt gp120, gp120 with the signal peptide of HA (HA-gp120), wild-type HA (HA-HA) and HA with the signal peptide of gp120 (gp120-HA) were labeled for 5 min and chased for indicated times. Ru: reduced gp120 with the signal peptide still attached; Rc: signal-peptide cleaved gp120; wt: wild-type gp120. Gels shown are representative of at least three independent experiments performed with fresh cells and transfections (biological replicates).
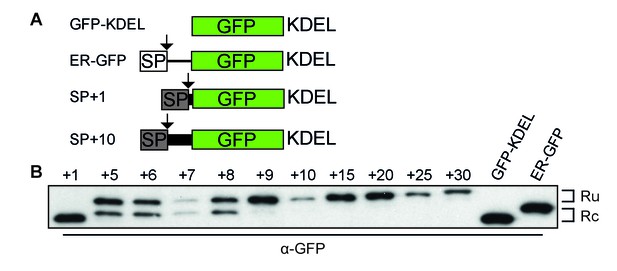
Downstream residues of gp160 regulate signal-peptide cleavage.
(A) Schematic of different GFP reporter constructs generated for imaging and immunoblot experiments. All reporters have the identical monomeric GFP-KDEL cassette. For ER GFP, the GFP-KDEL cassette is preceded by the bovine prolactin signal peptide (open box SP). The reporters with the HIV Env signal peptide (grey box SP) are fused to one or more amino acids of the mature gp120 domain followed by the GFP-KDEL cassette. (B) Western Blot analysis of control constructs (GFP-KDEL, ER-GFP) or gp160 signal peptide with 1-30 downstream residues (SP +1, …, 30). GFP-KDEL and SP +1 run lower on gel as they lack the additional residues downstream of the cleavage site. Ru: unprocessed molecules with signal peptide still attached; Rc: signal peptide-cleaved molecules. All images shown are representative of at least two independent experiments performed with fresh cells and transfections (biological replicates).
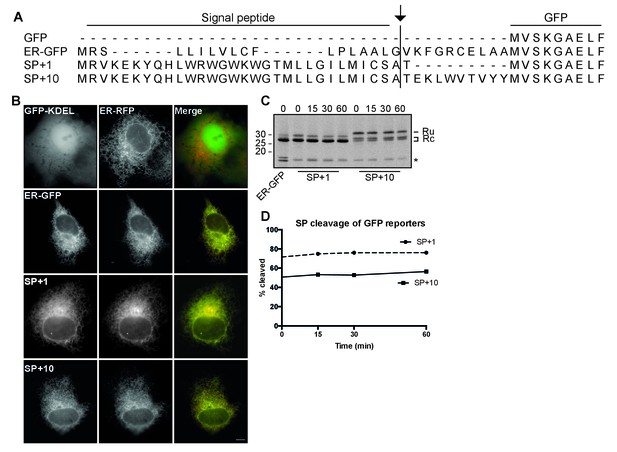
Characterization of SP-GFP fusion constructs.
(A) Sequence alignment of GFP-fusion constructs. Protein-sequence alignment of GFP-gp120 fusion constructs used in Figure 2. Only the first 9 residues of GFP are included for brevity. The arrow indicates the predicted signal-peptide cleavage site. All constructs contain a C-terminal ER-retrieval signal KDEL. (B) Fluorescence microscopy of Cos-7 cells expressing GFP constructs containing ER-retention signal KDEL. GFP-KDEL: cytosolic control GFP; ER-GFP: ER-resident GFP with the signal peptide of hen-egg lysozyme; SP +1: GFP with gp160 signal peptide and one gp160 residue as linker; SP +10: GFP with gp160 signal peptide and ten gp160 residues; ER-RFP: ER marker. Scale bar = 10 μm. (C) ER-GFP, SP +1 and SP +10 were analyzed by pulse chase as in Figure 1A. (D) Quantifications from C. *: background band.
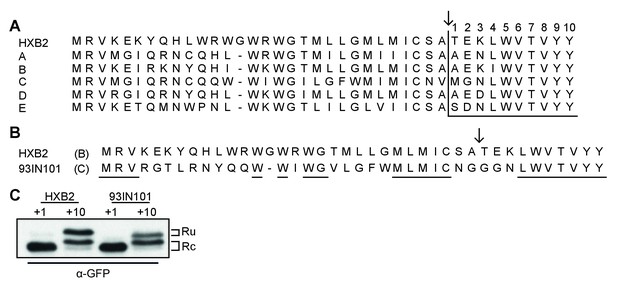
Env posttranslational signal-peptide cleavage is conserved between subtypes.
(A) Alignment of the signal peptide of HIV-1 Env reference strain HXB2 and the consensus sequences for subtypes A-E (www.hiv.lanl.gov). (B) Sequence alignment of HXB2 and subtype C strain 93IN101. Residues underlined are conserved between the two strains. (C) Western Blot analysis of HXB2 and 93IN101 SP +1 GFP and SP +10 GFP reporters. Blot shown is representative of two independent experiments performed with fresh cells and transfections (biological replicates).
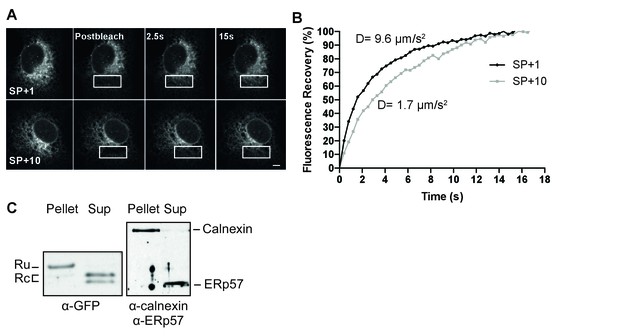
The uncleaved signal peptide acts as a signal anchor.
(A) FRAP analysis of gp160 signal-peptide constructs as in Figure 2. Cos-7 cells expressing SP+1 and SP+10 GFP reporters were subjected to FRAP analysis. A small region of interest (white outlined box) was photobleached with intense laser light and imaged with low laser light to visually (A) and quantitatively (B) compare mobilities and fluorescence-intensity recovery rates. (A) Both reporters are mobile and unbleached reporters diffuse into the photobleached region of interest. Scale bar = 10 μm. (B) Plot of representative fluorescence recoveries into the photobleach region of interest reveals that SP+10 is slower to recover. Number of cells analyzed, diffusion constants and, statistical values can be found in Figure 4—source data 1. (C) Western Blot analysis of pellet and supernatant (sup) fractions from a carbonate extraction of cells expressing SP+10. Split band for GFP in Sup is likely due to fragmentation (Wei et al., 2015). Blots in panel C are representative of at least two independent experiments (biological replicates).
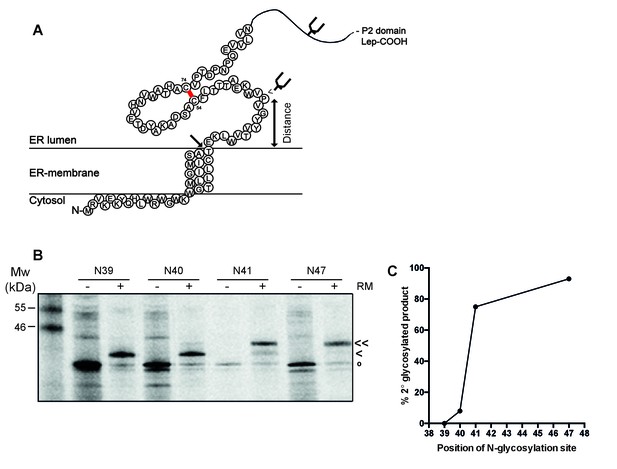
The signal-peptide cleavage site is buried in the membrane.
(A) Cartoon of the first 89 residues of gp160 attached to the P2 domain of protein leader peptidase (Lep) at the C-terminus. A first glycosylation site in Lep was used as translocation control. The second site was introduced at positions 39–47 to determine at which position the distance to the membrane is sufficient for glycosylation. Arrow indicates site of signal-peptide cleavage. (B) Gp160-Lep constructs with glycosylation sites N39, N40, N41, and N47, were in-vitro translated in presence (+) or absence (-) of dog pancreas microsomes (RM). Gp160-Lep received a single (<) or double (<<) glycan, or remained unglycosylated/untranslocated (o). (C) Band intensities of B were quantified and percentage of second glycosylation product was plotted against position of the engineered glycosylation site. The data in panels B and C are representative of multiple independent experiments (biological replicates).
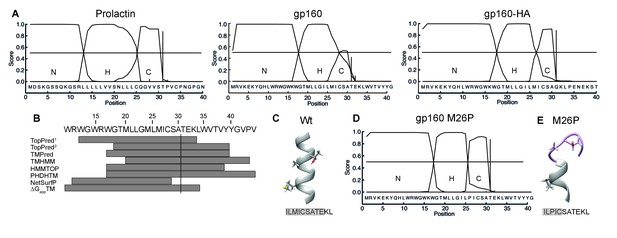
The C region of the gp160 signal peptide overlaps with the hydrophobic core region.
(A) Signal-peptide prediction tool SignalP 3.0 (36) was used to assess signal peptides of prolactin, gp160, and gp160 signal peptide followed by HA (as used in Figure 1B). The characteristic N-terminal charged region (N), the hydrophobic membrane-spanning region (H), and the C-terminal region (C) containing the cleavage site were plotted. Vertical bars represent the first amino acid after the cleavage site. (B) TopPred1 (Goldman, Engelman, and Steitz scale), TopPred2 (Kyte and Doolittle scale), TMPred, TMHMM, HMMTOP, PHDHTM, and NetSurf Helix were used to predict the transmembrane domain of the gp160 signal peptide. The predicted transmembrane domains are represented by grey bars below the HXB2 sequence (residues 12–45). The cleavage site is marked by a vertical line. (C) Robetta (Kim et al., 2004) was used to predict the structure of the area around the signal-peptide cleavage site of wild-type gp160. Residues M26, A30 and T31 are shown as sticks. Alpha helices colored in grey. (D) SignalP prediction of gp160 M26P signal peptide. (E) as in (C) except M26P gp160 was used for the prediction. Structures shown are representative of the 5 predicted structures received from the Robetta server.
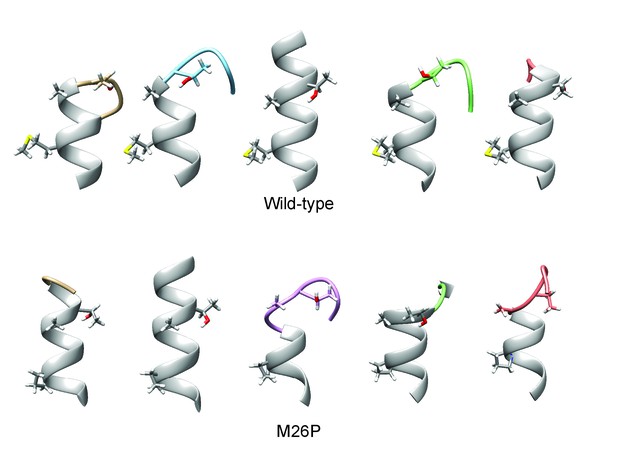
De-novo structure predictions of gp160 wild-type and M26P signal peptide.
As in Figure 6C and E. All predicted structures are shown, Central (purple) predictions were included in Figure 6.
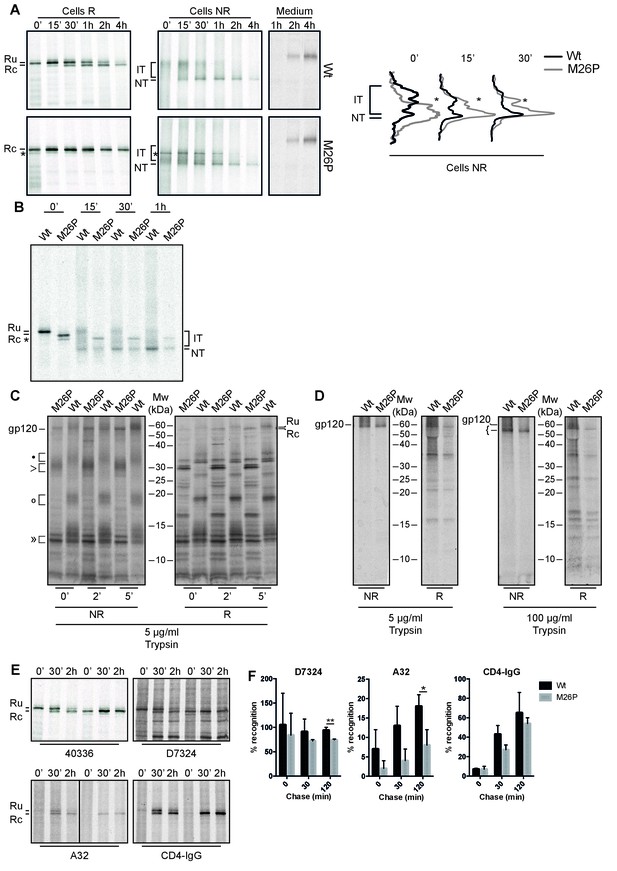
M26P leads to co-translational signal-peptide cleavage.
Experiments were done as in Figure 1. (A) HeLa cells expressing wild-type and M26P gp160 were radiolabeled for 10 min and chased for the indicated times. Samples were deglycosylated with endoH and subjected to reducing (Cells R) and non-reducing (Cells NR) 7.5% SDS-PAGE. Medium samples were reduced and not deglycosylated. Lane profiles depicting the folding-intermediate (NR) smear of wild-type and M26P gp160 were determined from autoradiographs. (B) As in A except that wild-type and M26P gp120 were used and samples were pulse labeled for 5 min in the presence of 5 mM DTT and chased in the absence of DTT. (C + D) HeLa cells expressing wild-type and M26P gp120 were pulse labeled as above and chased for either 0, 2, or 5 min (C) or 2 h (D). At the end of each time point, detergent cell lysates were proteolyzed with 5 or 100 µg/ml trypsin for exactly 15 min on ice. Proteolyzed samples were processed as in Figure 1 and analyzed by 15% SDS-PAGE. (E) HeLa cells expressing wild-type or M26P gp160 were pulse labeled and chased as above. Detergent cell lysates were immunoprecipitated in parallel with either polyclonal antibody 40336 or antibodies A32, D7324, or CD4-IgG. After immunoprecipitation, samples were processed as in Figure 1. (F) Quantifications of experiments from E. Values were normalized compared to immunoprecipitation by 40336. Statistics were calculated using an unpaired t-test with Welch’s correction. Exact p values can be found in Figure 7—source data 1. IT: folding intermediates; NT: native gp160; *: uncleaved unglycosylated M26P gp160 that had not targeted properly to the ER, likely due to its suboptimal signal sequence. Gels shown are representative of at least 3 independent experiments (biological replicates).
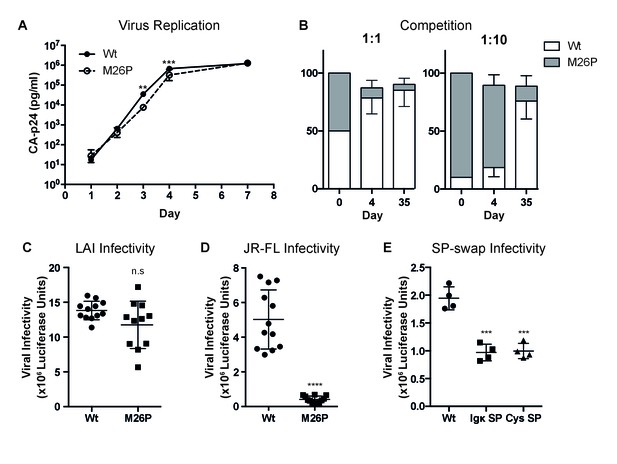
Co-translational signal-peptide cleavage causes functional gp160 defects.
(A) SupT1 cells (5x106) were infected with 20 µg of wild-type or M26P-mutant pLAI DNA constructs using electroporation. Replication was monitored by CA-p24 ELISA. The replication curves shown represent the averaged values of 6 independent experiments. p=0.0058 (**) and 0.003 (***). (B) SupT1 cells (Krogh et al., 2001) were infected with a total of 50 pg CA-p24 equivalent wild-type or M26P-mutant LAI virus (produced by transfected HEK293T cells) in ratios 1:1 and 1:10, each in duplicate (biological replicate). After 4 and 35 days, the isolated virus from each culture was sequenced and the electropherograms were quantified. (C,D,E) TZM-bl reporter cells were infected with: 500 pg of wild-type or M26P LAI virus (produced by HEK293T cells) (n.s, p=0.0833) (C), 1,000 pg of wild-type or M26P-mutant JR-FL pseudo-virus (produced by HEK293T cells) (p<0.0001). One M26P data point was excluded as it lay well below the background. (D), 1,000 pg of wild-type, Ig κ (p=0.0004) or cystatin-SP (p=0.0005) mutant LAI pseudo-virus (produced by C33A cells) (E) and infectivity was measured by Luciferase activity. The data in panels A, C and D are derived from 3 independent experiments, using 3 independently produced (pseudo-)virus stocks (biological replicates), each performed in quadruplicate (technical replicates). The data in panel B are derived from 2 independent experiments, using 2 independently produced virus stocks (biological replicates) and sequenced with two independent primers (technical replicates). The data in panel E are derived from 1 pseudovirus stock (biological replicate), performed in quadruplicate (technical replicates). All significance values were calculated with an unpaired, two-tailed t test with Welch’s correction.
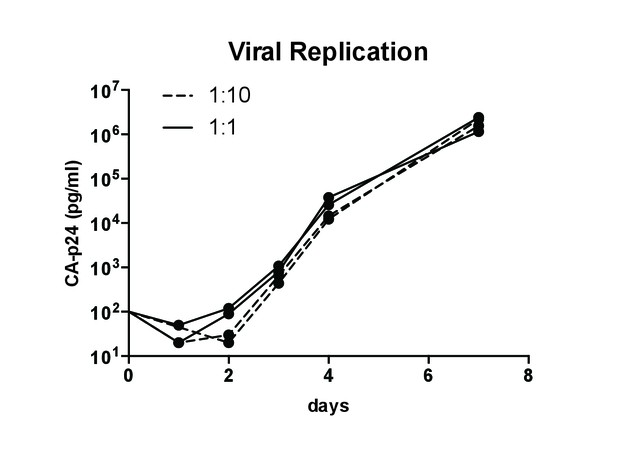
Viral replication in competition assays.
SupT1 cells (105) were infected with a total of 50 pg wild-type and M26P mutant LAI virus (produced on HEK293T cells) in ratio 1:1 (solid line) and 1:10 (dashed line) as in Figure 8B. Replication was monitored by CA-p24 ELISA. Each line shows replication of an independent culture (biological replicate).
Tables
M26 and I26 have highest conservation in gp160 sequences An alignment of 4236 gp160 sequences (Supplementary file 1) was used to compare absolute occurrence of each amino acid in position 26 of the signal peptide (SP) and the respective cleavable SP probability predicted by SignalP 3.0.
https://doi.org/10.7554/eLife.26067.015| Position 26 | SP probability | Absolute occurrence |
|---|---|---|
| Y | 0.458 | 0 |
| I | 0.51 | 1375 |
| F | 0.591 | 7 |
| V | 0.604 | 16 |
| M | 0.628 | 2638 |
| L | 0.697 | 143 |
| C | 0.699 | 1 |
| W | 0.751 | 1 |
| T | 0.777 | 39 |
| G | 0.792 | 0 |
| A | 0.805 | 2 |
| S | 0.848 | 5 |
| K | 0.863 | 0 |
| Q | 0.897 | 0 |
| E | 0.901 | 4 |
| D | 0.906 | 0 |
| R | 0.909 | 3 |
| N | 0.923 | 0 |
| H | 0.928 | 1 |
| P | 0.928 | 1 |
Additional files
-
Supplementary file 1
Amino acid and N-linked glycan conservation across gp160 sequences.
The 2014 filtered alignment of >4100 gp160 sequences was downloaded from the Los Alamos National Laboratory HIV-1 molecular database (www.hiv.lanl.gov) and placed into Microsoft Excel to calculate amino acid conservation at each position.
- https://doi.org/10.7554/eLife.26067.016





