Metaphase chromosome structure is dynamically maintained by condensin I-directed DNA (de)catenation
Figures
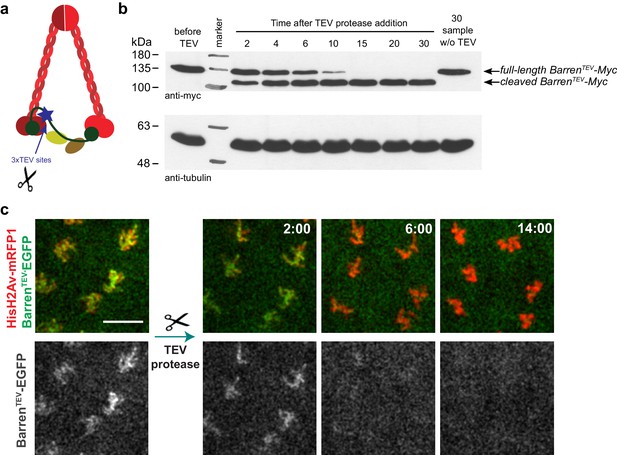
TEV-mediated cleavage of Barren disrupts condensin I function within a few minutes.
(a) Schematic representation of condensin complex indicating the position of the 3xTEV cleavage sites in the kleisin subunit Barren (aa175). (b) In vitro cleavage of BarrenTEV-myc. Extracts were prepared from ovaries of flies expressing solely TEV-cleavable Barren and incubated with TEV protease for the indicated time points (periods of time). The presence of full-length and cleaved Barren was monitored by western blot using myc antibodies. Tubulin was used as loading control. (c) Early embryos (0–30 min old) expressing HisH2AvD-mRFP1 (red) were injected with mRNA coding for BarrenTEV-EGFP (green). Embryos were aged for 1 hr-1hr 30m to allow for protein expression. Embryos were injected with 12 mg/ml UbcH10C114S protein to arrest in metaphase and subsequently with TEV-protease; images depict the same region before and after TEV injection; times (minutes:seconds) are relative to the time of injection; scale bar is 10 μm.
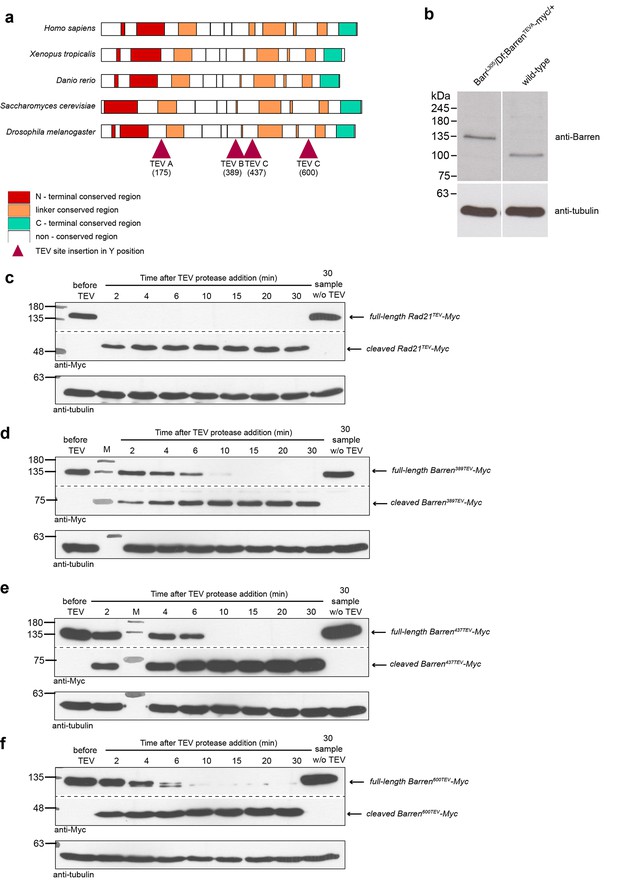
A TEV-cleavable system to destroy condensin I.
(a) graphic representation of the four positions used to introduce three consecutive TEV-protease consensus sites within the linker region of Drosophila Barren. Conservation with other species is shown and conserved regions are colour-coded. (b) Western blot analysis of the strain carrying solely Barren175TEV-myc, compared to wild-type strains, confirming the absence of endogenous protein. Extracts were prepared from ovaries and probed using the indicated antibodies. (c–f) Western blot analysis of in vitro cleavage of different versions of myc-tagged BarrenTEV or Rad21TEV. Extracts were prepared from ovaries of flies expressing TEV-cleavable Barren/Rad21 and incubated with TEV protease for the indicated time points. The presence of full-length and cleaved Barren was monitored by western blot using myc antibodies. Tubulin was used as loading control.
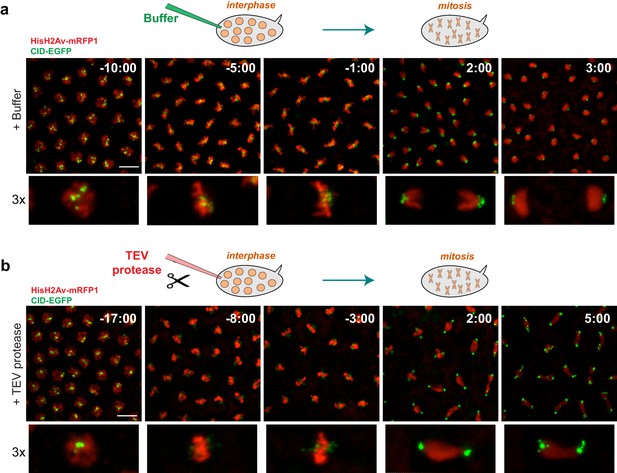
Condensin I inactivation prior to mitotic entry.
Embryos surviving solely on BarrenTEV were injected with buffer (a) or 13 mg/ml TEV protease (b) ~10–15 min before mitosis; Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Bottom rows show higher magnifications (~3x) of a single nuclear division. Times (minutes:seconds) are relative to the time of anaphase onset.
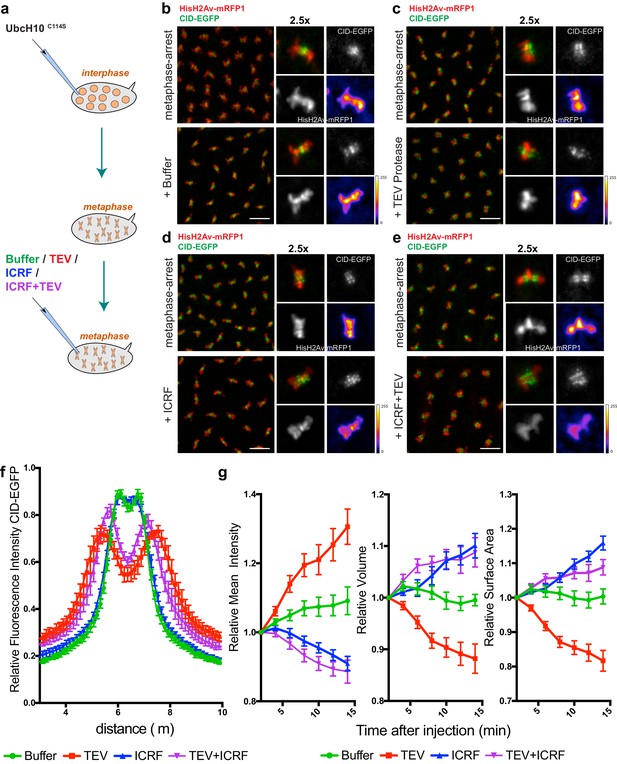
Condensin I inactivation in pre-assembled chromosomes leads to disruption of centromere structure and hyper-compaction of mitotic chromosomes.
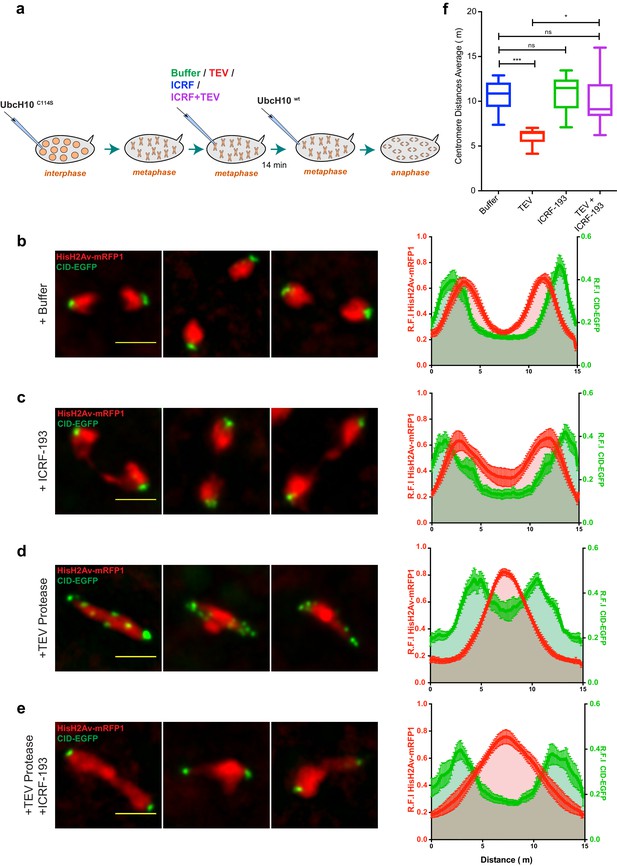
(a) Schematic representation of the experimental layout. Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S) to induce a metaphase arrest. Embryos were subsequently injected with buffer (b), 13 mg/ml TEV protease (c), 280 μM ICRF (d) or a mixture containing 13 mg/ml TEV protease and 280 μM ICRF (e); Images depict embryos before the second injection and 14 min after. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Insets show higher magnifications (2.5x) of a single metaphase. Times (minutes:seconds) are relative to the time of the second injection. (f) Quantitative analysis of centromere positioning 10 min after the second injection, as above; graph shows average ± SEM of individual embryos (n ≥ 7 embryos for each experimental condition); for each embryo, a minimum of 8 metaphases was measured; (g) quantifications of mean voxel intensity, volume and surface area of the entire metaphase plate quantified in 3D, over time, and normalized to the time of the second injection. Graphs represent the average ± SEM of individual embryos (n ≥ 10 embryos for each experimental condition); for each embryo, a minimum of 8 metaphases was quantified.
-
Figure 3—source data 1
Centromere displacement and chromosome compaction measurements upon condensin I and topoII inactivation.
- https://doi.org/10.7554/eLife.26120.009
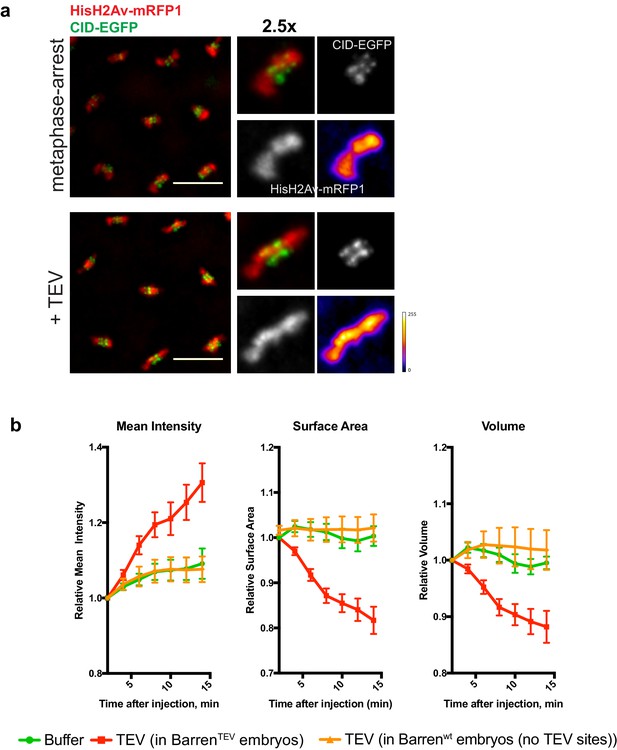
- Chromosome condensation induced by TEV-protease depends on TEV cleavage sites present in BarrenTEV.
(a) Representative images from embryos that do not contain TEV-cleavage sites in Barren. Embryos were injected with UbcH10C114S to induce a metaphase arrest and subsequently injected with 13 mg/ml TEV protease. Embryos also express HisH2Av-RFP1 (red) and Cid-EGFP (green); scale bar is 10 μm. (b) Quantifications of mean voxel intensity, volume and surface area of the entire metaphase plate quantified in 3D, over time, and normalized to the time of the second injection. Graphs represent the average ± SEM of individual embryos (n ≥ 10 embryos for each experimental condition); for each embryo, a minimum of 8 metaphases was quantified.
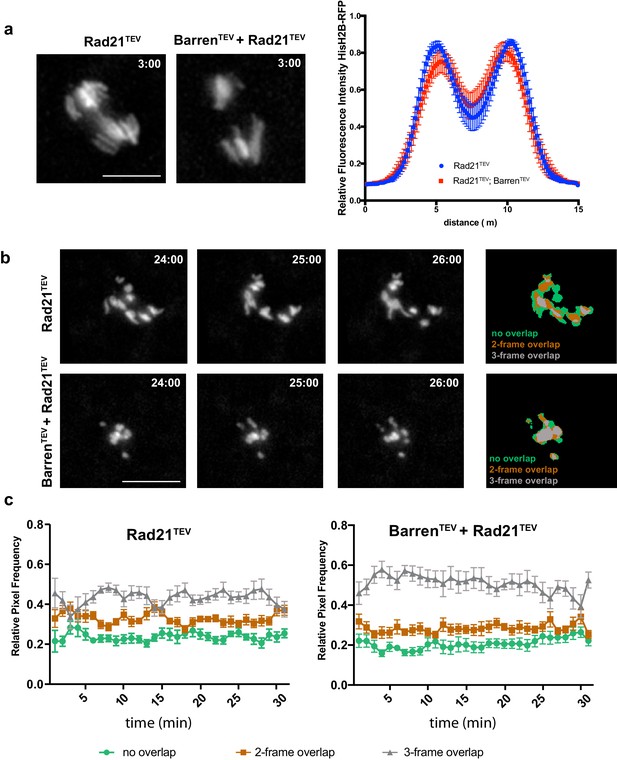
Condensin I inactivation in separated sister chromatids reduces their movement.
(a) Representative images of the initial separation after TEV-mediated cleavage of Rad21TEV and Rad21TEV + BarrenTEV. Graph plots the relative distribution of HisH2B-RFP at the maximal state of sister chromatid separation triggered by TEV-mediated cleavage of Rad21TEV, in strains that contain solely Rad21TEV or both Rad21TEV and BarrenTEV. A 15 μm line was used to measure plot profiles along the segregation plane, measured 3–5 min after TEV protease injection. Graphs plot the average ± SEM of individual embryos (n ≥ 7 embryos for each experimental condition). For each embryo, between 8 and 12 anaphases were analysed. (b) Example of chromosome movement analysis; left panel represents average of the binary images of three consecutive frames, used to estimate chromosome displacements: blue, non-overlapping pixels; green, two- out of three-frame overlap; grey, three-frame overlap. Scale bar is 10 μm. (c) Frequency of overlapping pixels to estimate chromosome displacement (as in b), over time, after TEV protease injection.
-
Figure 4—source data 1
Measurements of segregation efficiency and chromosome movement upon cohesin/condensin inactivation.
- https://doi.org/10.7554/eLife.26120.016
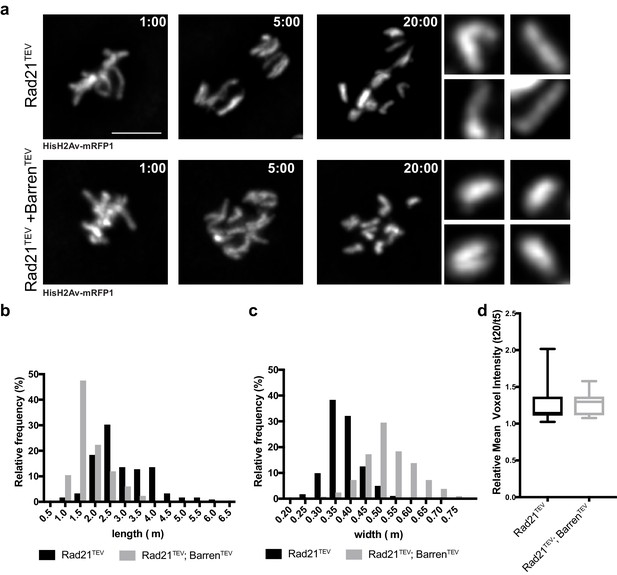
Chromosome over-compaction depends on sister-chromatid proximity.
(a) Stills from metaphase-arrested embryos after injection of TEV protease in strains surviving solely on Rad21TEV (cohesin cleavage) or Rad21TEV+BarrenTEV (cohesin and condensin cleavage); embryos also express HisH2B-RFP; scale bars, 5 μm. Insets show higher magnifications (3x) of single chromatids 20 min after TEV injection. Times (minutes:seconds) are relative to the time of TEV injection. (b–c) Relative frequency of sister chromatid length (b) and width (c) at 20 min after TEV injections (n ≥ 120 single chromatids from seven independent embryos for each experimental condition). (d) Mean voxel intensity of isolated single chromatids 20 min after TEV injections, normalized to mean voxel intensity 5 min past injection. (n ≥ 10 embryos for each experimental condition).
-
Figure 5—source data 1
Measurements of isolated chromatids upon cohesin/condensin inactivation.
Individual measurements of chromosome thickness, length and mean voxel intensity upon TEV-mediated cleavage of Read21TEV and Rad21TEV+BarrenTEV. Each data set is presented on a separate sheet. File includes descriptive statistics.
- https://doi.org/10.7554/eLife.26120.018
Condensin I inactivation results in TopoII-dependent sister chromatids intertwines and segregation failure.
(a) Schematic representation of the experimental set-up. Embryos were arrested with 12 mg/ml UbcH10C114S and injected with buffer (b), 280 μM ICRF-193 (c), 13 mg/ml TEV protease (d) or TEV+ICRF-193, while in metaphase; Embryos were subsequently injected with 14 mg/ml of a wild-type version of UbcH10 to release them from the arrest. Images depict representative images of the anaphase; Graphs plot the relative distribution of HisH2Av-mRFP1 and Cid-EGFP across the 15 μm segregation plane, measured 4–6 min after anaphase onset. Graphs plot the average +_SEM of individual embryos (n ≥ 10 embryos for each experimental condition). For each embryo, at least eight anaphases were analysed. (f) Quantification of centromere distances during UbcH10wt-induced anaphase as in (b–e). Graphs plot the distances between segregating centromeres measured 6 min after anaphase onset (n ≥ 10 embryos for each experimental condition; for each embryo, at least eight anaphases were analysed). Statistical analysis was performed using the non-parametric Kruskal-Wallis test; ns p>0.05, *p<0.05; ***p<0.001.
-
Figure 6—source data 1
Measurements of segregation efficiency after metaphase-specific inactivation of condensin and/or Topoisomerase II.
Anaphase profiles for HisH2Av-mRFP1 and Cid-EGFP measured 4–6 min after anaphase onset (Figure 6b-2). Each measurement represents the average for independent embryos (resulting from at least eight anaphases measured). Individual sheets include either the same measurement for the four experimental conditions or both Cid-EGFP and HisH2Av-mRFP1 for the same experiment, as indicated. Centromere separation measurements (Figure 6f) also include descriptive statistics.
- https://doi.org/10.7554/eLife.26120.022
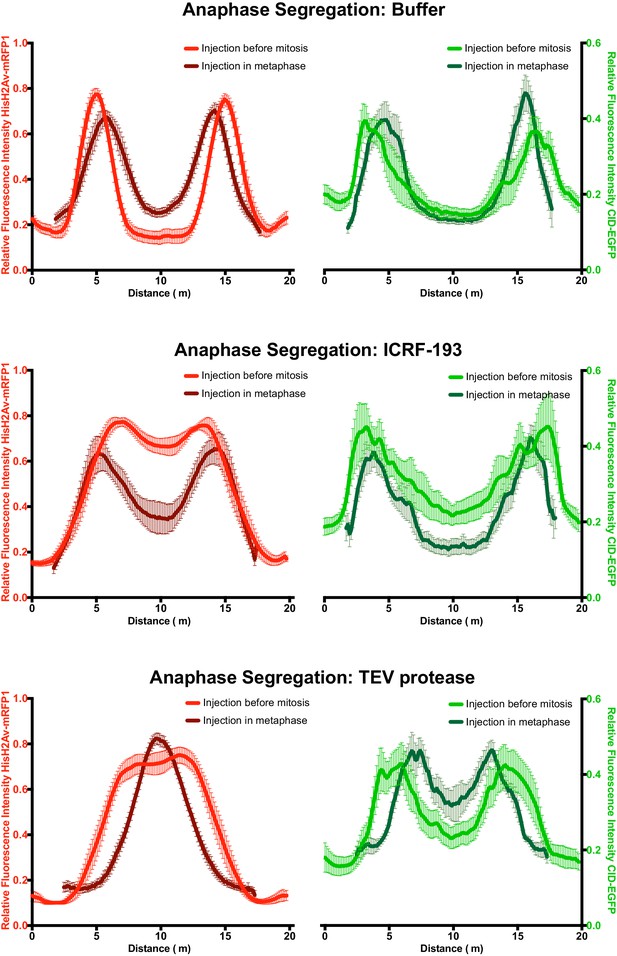
– Comparative analysis of segregation efficiency for condensin and/TopoII inhibition before mitosis (light colour) and during metaphase arrest/release (dark colour); Graphs plot the relative distribution of HisH2Av-mRFP1 (red) and Cid-EGFP (green) across a 20 μm segregation plane, measured 4–6 min after anaphase onset.
Graphs plot the average +_SEM of individual embryos (n ≥ 8 embryos for each experimental condition). For each embryo, at least eight anaphases were analysed.
Videos
Mitosis in Drosophila embryos.
Embryos were injected with buffer in early interphase and monitored throughout the subsequent mitosis. Embryos express HisH2Av-mRFP1 (red) and Cid-EGFP (green). Times are relative to injection time. Scale bar is 10 um.
Mitosis upon condensin I inactivation in Drosophila embryos.
Embryos surviving solely on BarrenTEV were injected with TEV protease in early interphase and monitored in the subsequent mitosis. Embryos express HisH2Av-mRFP1 (red) and Cid-EGFP (green). Times are relative to injection time. Scale bar is 10 um.
Buffer injection in metaphase-arrested embryos.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with buffer. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Times (minutes:seconds) are relative to the time of buffer injection.
Condensin I inactivation in metaphase-arrested embryos.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 13 mg/ml TEV protease. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Times (minutes:seconds) are relative to the time of TEV injection.
Topoisomerase II inhibition in metaphase-arrested embryos.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 280 μM ICRF-193. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Times (minutes:seconds) are relative to the time of ICRF injection.
Concomitant inactivation of Topoisomerase II and Condenin I in metaphase-arrested embryos.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with a mix of 280 μM ICRF-193 and 13 mg/ml TEV protease. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm. Times (minutes:seconds) are relative to the time of the second injection.
Artificial induction of sister chromatid separation in metaphase-arrested embryos.
Embryos expressing solely Rad21TEV and wild-type Barren were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 13 mg/ml TEV protease. Embryos also express His2B–RFP; scale bars, 10 μm. Times (minutes:seconds) are relative to the time of the second injection.
Effect of condensin I inactivation on isolated sister chromatids.
Embryos expressing uniquely TEV-sensitive Rad21 and Barren were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 13 mg/ml TEV protease. Embryos also express His2B–RFP; scale bars, 10 μm. Times (minutes:seconds) are relative to the time of the second injection.
Induced anaphase in control embryos.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with buffer. After 14 min embryos were injected a wild-type version of UbcH10 to induce anaphase. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm.
Induced anaphase after timely inhibition of topoisomerase II.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 280 μM ICRF-193. After 14 min embryos were injected a wild-type version of UbcH10 to induce anaphase. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm.
Induced anaphase after timely inhibition of Condensin I.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with 13 mg/ml TEV protease. After 14 min embryos were injected a wild-type version of UbcH10 to induce anaphase. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm.
Induced anaphase after timely inhibition of Condensin I and topoisomerase II.
Embryos expressing solely BarrenTEV were injected with 12 mg/ml of a dominant-negative form of the human E2 ubiquitin-conjugating enzyme (UbcH10C114S), to induce a metaphase arrest, and subsequently injected with a mix of 280 μM ICRF-193 and 13 mg/ml TEV protease. After 14 min embryos were injected a wild-type version of UbcH10 to induce anaphase. Embryos also express His2A–mRFP1 (red) and Cid-EGFP (green); scale bars, 10 μm.
Tables
List of fly strains used in this study
| CHR#* | Genotype | Reference |
|---|---|---|
| 1418 | BarrL305/CyO | Bhat et al. (1996) (RRID:BDSC_4402) |
| 1421 | Df(2L)Exel7077/CyO | Blommington #7850 (RRID:BDSC_7850) |
| 1513 | w;; Barr(175 - 3TEV)-myc10 III.5 | This study |
| 1509 | w; Barr(175 - 3TEV)-myc10 II.1; | This study |
| 1522 | w;; Barr(389 - 3TEV)-myc10 III.2 | This study |
| 1514 | w;; Barr(437 - 3TEV)-myc10 III.1 | This study |
| 1520 | w;; Barr(600 - 3TEV)-myc10 III.3 | This study |
| 1525 | w;; Barr(wt)-myc10 III.1 | This study |
| 1560 | w; BarrL305/ Df(2L)Exel7077; Barr(175 - 3TEV)-myc10 III.5 | This study |
| 820 | w;; HisH2AvD-mRFP1 III.1, CGC (CID-EGFP) III.1 | Schuh et al. (2007) |
| 1564 | Df(2L)Exel7077 / CyO; HisH2AvD-mRFP1 III.1, CGC (CID-EGFP) III.1 | This study |
| w; BarrL305/ Df(2L)Exel7077; Barr(175 - 3TEV)-myc10 III.5/ HisH2AvD-mRFP1 III.1, CGC (CID-EGFP) III.1 | ||
| 629 | w;; Rad21ex15, polyubiq-H2B-RFP, tubpr-Rad21(550-3TEV) -myc10 | Oliveira et al. (2010) |
| 1646 | w; BarrL305, Barr(175 - 3TEV)-myc10 II.1; +/+ | This study |
| 1648 | w; BarrL305, Barr(175 - 3TEV)-myc10 II.1; Rad21ex15, polyubiq-H2B-RFP, tubpr-Rad21(550-3TEV) -myc10 | This study |
-
*Reference number in our internal lab fly database





