A potent human neutralizing antibody Fc-dependently reduces established HBV infections
Figures
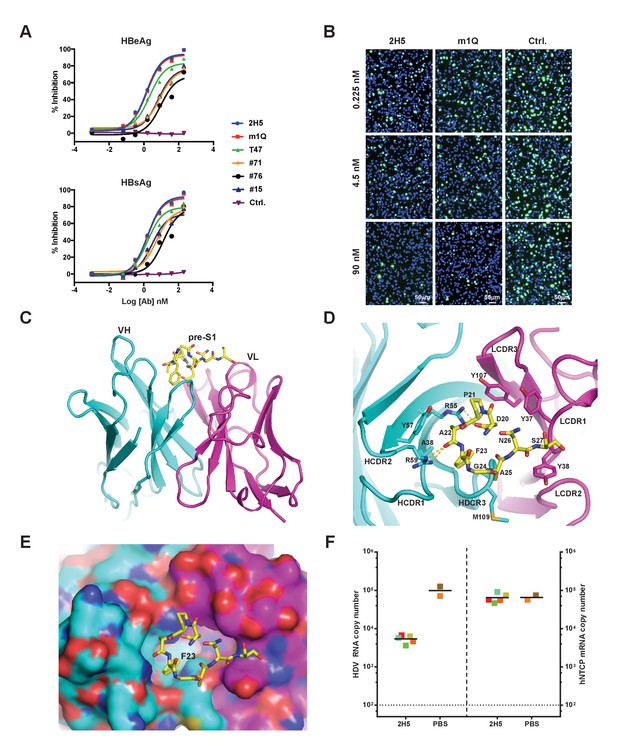
Identification, in vivo activity verification, and structural characterization of 2H5.
(A) Neutralization of HBV (genotype D) infection in the HepG2-hNTCP stable cell line by anti-preS1 Abs. Cells were infected with 200 multiplicities of genome equivalents (mge) in the presence of the tested Abs. The secreted HBeAg and HBsAg levels in sample supernatants were measured at 7 days post infection (dpi). The HBV neutralization activity is here presented as the percentage of inhibition of secreted HBeAg and HBsAg, which was calculated by normalizing the ‘infection only’ reading to 0% inhibition. (B) Neutralization of HDV infection in HepG2-hNTCP cells by 2H5 and m1Q. The cells were infected with 500 mge of HDV in the presence of the test nAbs or matched isotype control Ab. At dpi 7, HDV delta antigens were stained with FITC-conjugated mAb, 4G5; the cell nuclei were stained with DAPI. (C) Crystal structure of the scFv of 2H5 in complex with the preS1 peptide. 2H5 is shown in ribbon and residues 20–27 of preS1 (genotype C) are shown in sticks. Carbon atoms are colored in cyan for VH of 2H5, magenta for VL of 2H5 and yellow for the preS1 peptide. Oxygen atoms are colored in red and nitrogen atoms are colored in blue. (D) Interaction between 2H5 and preS1. 2H5 is shown in ribbon and the preS1 epitope and selected CDR side chains are shown in sticks. This view is perpendicular to panel C. Dotted lines denote hydrogen bonds. (E) Surface view of the peptide binding pocket of 2H5. Same color-coding and orientation as panel D. (F) 2H5 protected hNTCP-Tg mice from HDV infection. Human NTCP-Tg homozygotes (C57BL/6 background) were IP administered 2H5 (20 mg/kg; n = 5) or PBS control (n = 2) at 8 days after birth. One hour later, each mouse was challenged with 1.47 × 1010 genome equivalents (GE) of HDV. The mice were sacrificed at dpi 6. HDV total RNA titers (on the left Y-axis) and hNTCP transgene expression (on the right Y-axis) in liver tissues were measured by qPCR. The horizontal dotted line indicates the reliable detection limit. The copy numbers shown in the Y-axes are from 20 ng of total liver RNA for each sample. Each square represents one mouse; squares of the same color indicate data from the same mouse.
-
Figure 1—source data 1
Data for Figure 1F.
2H5 protected hNTCP-Tg mice from HDV infection.
- https://doi.org/10.7554/eLife.26738.005

HBV envelope proteins and amino acid sequence alignment of preS1 peptides.
Schematic diagram of HBV envelope proteins:Small (S), middle (M) and Large (L) proteins. All three proteins share the same C-terminal S domain (226 aa). The L protein has an extra preS1 (108 aa or 119 aa depending on genotypes) and a preS2 (55 aa) domain. The L protein is myristoylated at the N-terminus. The N-terminal aa sequence of preS1 from different genotypes are shown. Two synthesized peptides marked with a star (m47b and NC36b) were used for phage display library selection. Both of these were derived from the preS1 domain of HBV genotype C and have a biotin modification at their C-termini. m47 is an N-terminal myristoylated peptide corresponding to aa 2–48 of the preS1 domain (m47). The other peptides shown were used for antibody binding activity assays and epitope mapping, and are based on HBV genotype C, with the exception of 47D, which is based on genotype D. The NTCP receptor binding site (RBS) of the preS1 peptide and the epitopes of four nAbs are indicated on the top of the alignment.

Binding of the six anti-preS1 nAbs to preS1 peptides and characterization of their epitopes.
(A) Binding of anti-preS1 nAbs to preS1 peptides by ELISA. The selected six Abs in their full-length human IgG1 forms were tested for binding to 188 nM preS1 peptides captured on an ELISA plate that had been pre-coated with streptavidin. 2H5, m1Q, and T47, bound to all three of the peptides. #71, and #76 bound to NC36b and 47b but not m47b. #15 bound to m47b and 47b, but not NC36b. The binding of Abs was detected by HRP-anti-human Fc secondary Ab. (B) ELISA-based assay of Abs binding to the shorter preS1 peptides, NN23b and LD23b. The assay was performed as in panel A. (C) Competition ELISA with short preS1 peptides. In the presence of competition short peptides without biotinylation at the indicated concentrations, the binding activity of Abs with 188 nM biotinylated m47b or 47b preS1 peptide was measured. (A–C) The different epitopes recognized by the nAbs are depicted in Figure 1—figure supplement 1. (D) Competition ELISA with mutant peptides. Amino acids of preS1, Leu19, Asp20, Pro21, or Phe23 were mutated to alanine in LN16 (a 16-mer pre S1 peptide) and tested for competition activity against the binding of 2H5 to m47b; the assay was performed as in panel C. (E) G24R mutation in preS1 did not affect 2H5 binding.
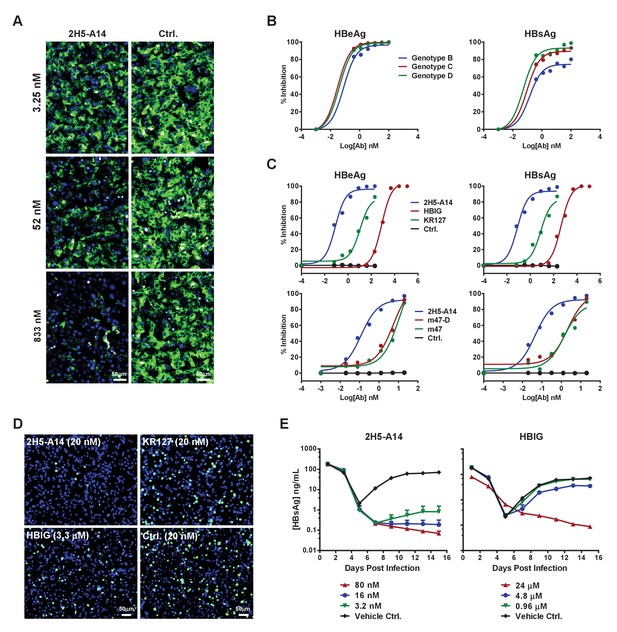
Blocking activity of 2H5-A14 for preS1 binding to hNTCP and the broad and potent neutralization activity of 2H5-A14 against HBV and HDV.
(A) 2H5-A14 competed for the binding of preS1 to HepG2-hNTCP cells. 2H5-A14 or an isotype-matched control Ab at the indicated concentrations and 100 nM of FITC-labeled preS1 lipopeptide were mixed, and added to HepG2-hNTCP cells and were incubated for 15 mins, followed by extensive washing and staining of nuclei by DAPI. (B–E) The broad and potent neutralization activity of 2H5-A14 against HBV and HDV infection. 2H5-A14 neutralized HBV genotypes B, C, and D (B). In comparison to the prototype preS1 lipopeptides of Myrcludex-B, KR127 and HBIG in neutralizing HBV infection of HepG2-hNTCP cells, 2H5-A14 showed superior activity (C). 2H5-A14 also potently neutralized HDV infection of HepG2-hNTCP cells (D) and HBV infection of PHHs (E). The neutralization assays were performed similarly as in Figure 1A–B.
-
Figure 2—source data 1
Data for Figure 2B, 2C and 2E.
The broad and potent neutralization activity of 2H5-A14 against HBV and HDV infection.
- https://doi.org/10.7554/eLife.26738.014
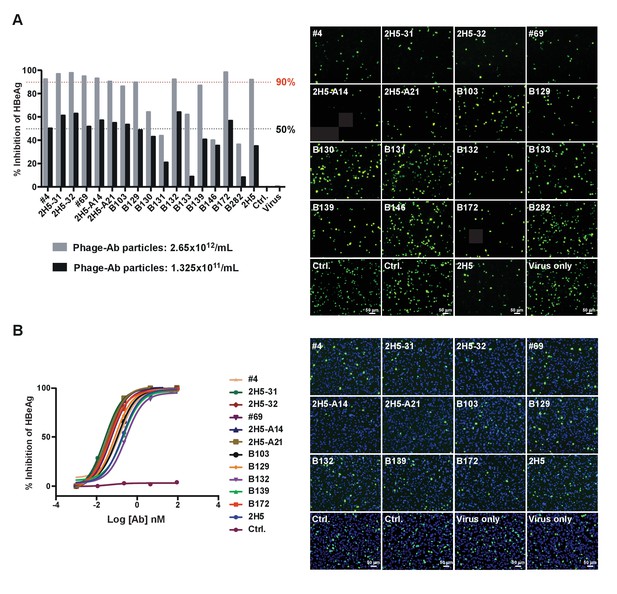
Neutralization of HBV and HDV infection by the 11 nAbs derived from 2H5-VH chain shuffling.
The neutralization assays were performed as in Figure 1. (A) The initial HBV and HDV neutralization screen using phage-scFv Abs. The phage-scFvs at the indicated concentrations were mixed with viruses and then added to HepG2-hNTCP cells to test for inhibition of viral infection. The percentage of HBeAg inhibition and the reduction of HDV Delta antigen staining (green) are shown at the left and the right, respectively. For HDV neutralization assay, the phage-scFv Abs were tested at 2.65 × 1012/mL. (B) Evaluation of the HBV (left) and HDV (right) neutralization activity of the 11 selected nAbs in their full-length hIgG1 forms. For HDV neutralization assay, the nAbs were tested at 1 nM.
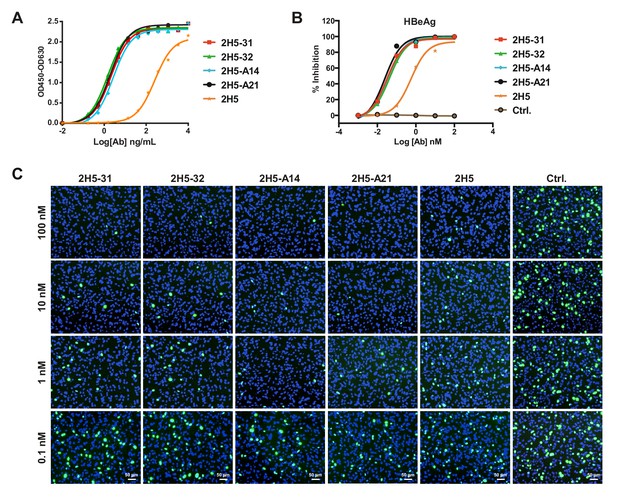
preS1 binding activity and neutralization activities of the top four nAbs obtained from 2H5-VH chain shuffling.
(A) Evaluating the binding activities of nAbs to preS1 peptide. The nAbs in their full-length human IgG1 forms were tested for binding to 7.8 nM NC36b peptides captured on an ELISA plate that had been pre-coated with streptavidin. (B-C) Neutralization of HBV and HDV by nAbs. The percentage of HBeAg inhibition at dpi seven is shown in panel (B); the staining of HDV delta antigens at seven dpi is shown in panel (C).
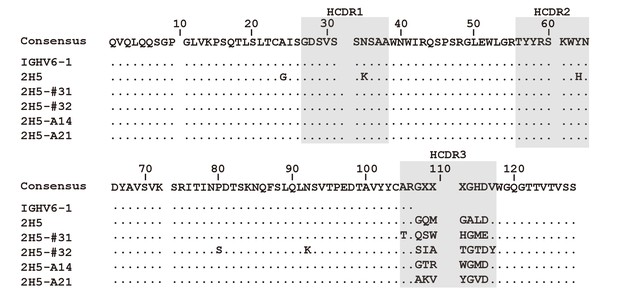
Amino acid sequence alignment of the VHs of nAbs.
The three CDRs of the VH were defined using the IMGT database, and are highlighted. The antibody numbering scheme used here was also based on the IMGT numbering system.

Epitope mapping of 2H5-A14.
(A) Competition ELISA. Short peptides or peptides containing a single amino acid change (as indicated) were used to compete for the binding activity of 2H5-A14 to biotinylated m47b (7.8 nM). The assay was performed similarly to that described in Figure 1—figure supplement 2C–D. (B) Binding of 2H5-A14 to a G24R mutation of preS1 (NC36b-G24R). 2H5-A14 at indicated concentrations were tested for binding to NC36b or NC36b-G24R peptides (7.8 nM) captured on an ELISA plate.
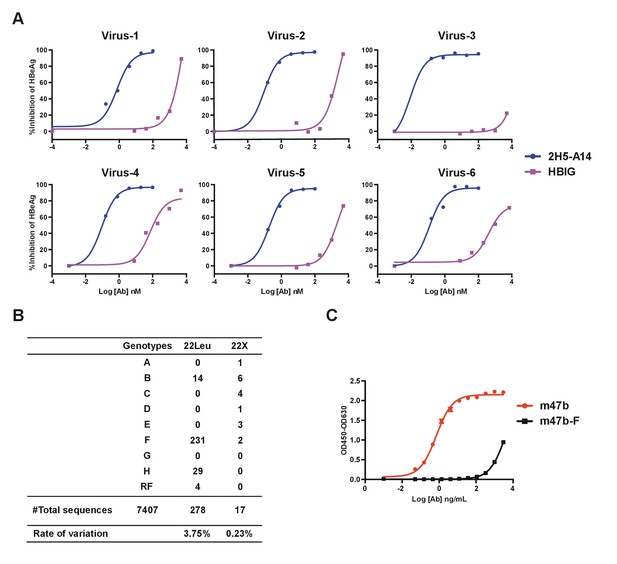
2H5-A14 neutralization of patient serum derived HBV viruses, and sequence variation analysis of the 2H5-A14 epitope among HBV genotypes.
https://doi.org/10.7554/eLife.26738.013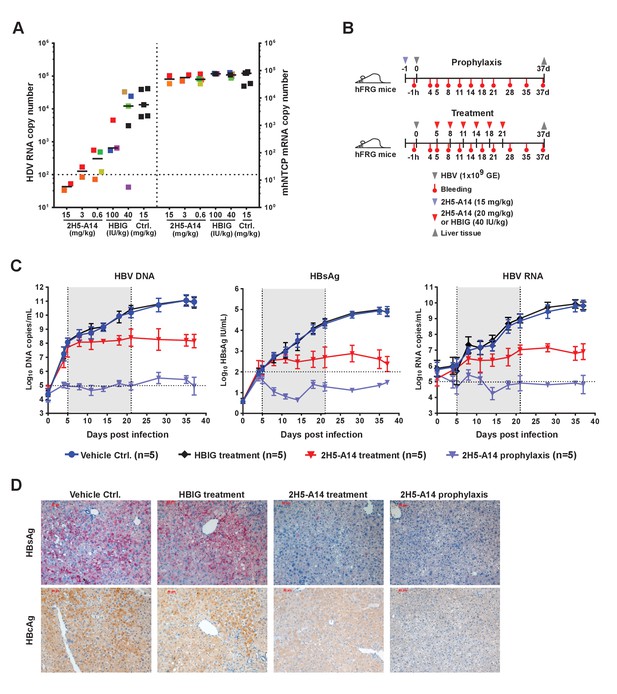
Prophylactic and therapeutic efficacy of 2H5-A14 in mouse models.
(A) 2H5-A14 protected NTCP-edited mice from HDV infection. A total of 21 mice (NTCP-edited homozygous FVB) were IP administered 2H5-A14, HBIG, or matched isotype control Ab at the indicated concentrations at 9 days after birth. One hour later, each mouse was challenged with 1 × 1010 GE of HDV and was then sacrificed on post infection day 6. The liver tissues were collected from each mouse. HDV total RNA levels and the edited NTCP gene expression were measured by qPCR. The data were presented similarly as in Figure 1F. (B) Schematic diagram illustrating HBV challenging, bleeding and antibody administration schedules in the mouse study. Five animals were used in each group. Recipient hFRG mice were challenged with 1 × 109 GE HBV (Genotype B) on day 0. A single dose of 15 mg/kg 2H5-A14 was IP administered one day before viral challenge (prophylaxis), or biweekly doses of 20 mg/kg 2H5-A14 were IP administered starting from day 5 to day 21 after the challenge (therapeutic). HBIG (40 IU/kg) treatment was used as a control. (C) Levels of HBV DNA and RNA, and HBsAg concentrations in mouse serum. Blood samples from all the mice (panel B) were collected at the indicated time points for measuring HBV DNA, HBV RNA, and HBsAg levels. The horizontal dotted lines indicate the reliable detection limits; the vertical dotted lines and the grey-shaded areas indicate the treatment window. (D) IHC staining of HBsAg and HBcAg in liver tissues from sacrificed mice at the end of the experiment. Intrahepatic HBsAg and HBcAg levels were detected by specific antibodies conjugated with Alkaline phosphatase (AP) and horseradish peroxidase (HRP), respectively. Each set of images represents the results for one mouse of each group.
-
Figure 3—source data 1
Data for Figure 3A and 3C.
- https://doi.org/10.7554/eLife.26738.019
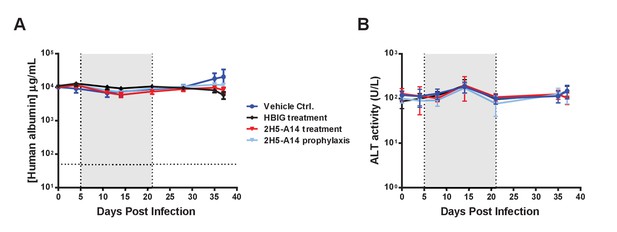
Human albumin level and ALT activity in hFRG mouse serum samples.
The mice were infected and treated as described in Figure 3B. Blood samples collected at the indicated time points were used for measuring human albumin level (A) and ALT activity (B) in mouse sera. The vertical dotted lines and the grey-shaded areas indicate the treatment window of HBIG or 2H5-A14. Human albumin levels at different time points post infection were quantified using a Human Serum Albumin ELISA Kit (Thermo Fisher). The horizontal dotted line indicates the reliable detection limit of human albumin. ALT activity was measured following the instructions of an ALT activity assay kit (Sigma).

Intrahepatic HBV total DNA, RNA, and cccDNA in liver tissues of HBV-infected hFRG mice.
Recipient mice were challenged with 1 × 109 GE HBV (genotype B) per mouse on day 1, followed by a single dose of 2H5-A14 IP either one day before viral challenge (prophylaxis) or by a biweekly IP administration of 2H5-A14 starting from day 5 to day 21 after the challenge (therapeutic). HBIG were included as a control group for the 2H5-A14 therapeutic group. All mice were sacrificed at 37dpi and liver tissues were collected. (A) Northern blotting of HBV total RNA in liver tissues. Total RNA samples were extracted and analyzed by northern blotting using an HBV-specific DIG-labeled probe. Total RNA (2 µg per lane) were loaded for each sample. The positions of progenomic RNA (3.5 kb) and subgenomic RNAs (2.4/2.1 kb) are indicated. (B) Southern blotting of HBV total DNA in liver tissues. The total DNA extracted from liver tissues were loaded at 500 ng per lane (one sample, one lane) and subjected to Southern blotting. HBV replication DNA intermediates were analyzed by Southern blotting using a DIG-labeled HBV DNA probe. (C) Southern blot analysis of cccDNA. HBV cccDNA was extracted from liver tissues using a Hirt protein-free method and was analyzed by Southern blotting.
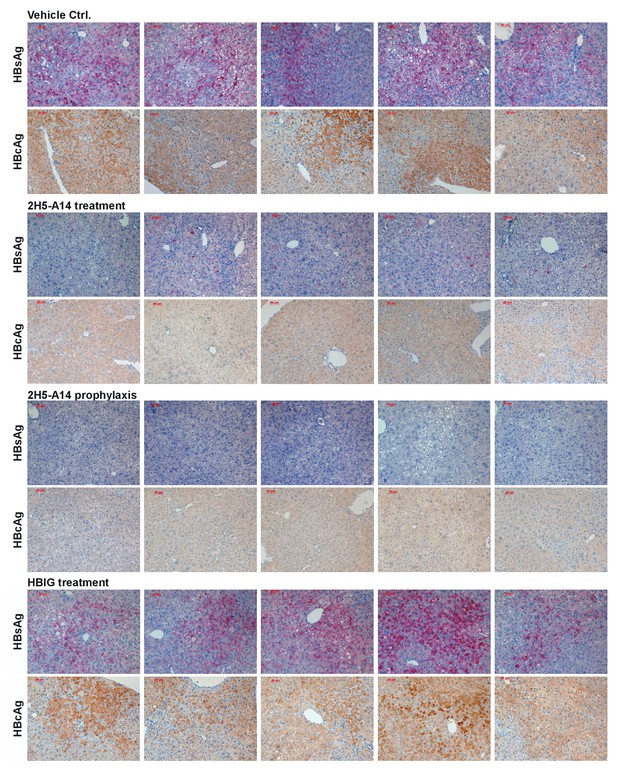
Intrahepatic HBsAg and HBcAg in liver tissues of HBV-infected hFRG mice.
The mice were infected and treated as described in Figure 3 and Figure 3—figure supplement 1. The HBsAg and HBcAg levels in liver tissues were detected by IHC. One set of images represents one mouse of each group. The representative images from each group shown in Figure 3D are also included in this figure as well to enable easier comparisons.
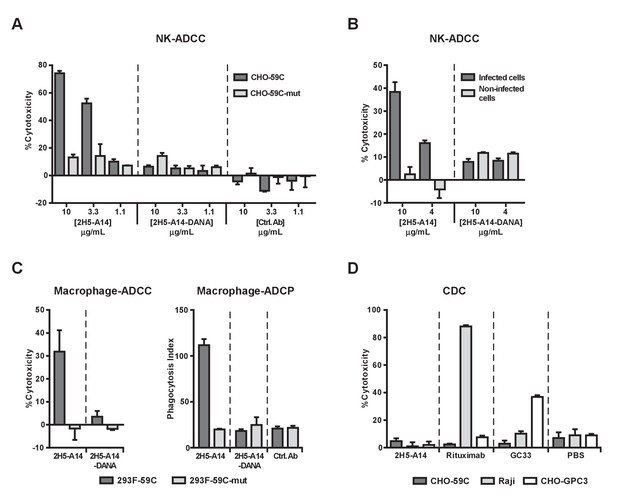
2H5-A14 mediated ADCC and ADCP but not CDC.
(A–B) 2H5-A14 induced ADCC via NK cells. NK92-MIhCD16 was used as effector cells. 2H5-A14, its ADCC-inactive Fc variant (2H5-A14-DANA) or isotype-matched control Abs were tested at the indicated concentrations with three replicates. Different target cells were analyzed, including CHO cells expressing the epitope of 2H5-A14 inserted into the extracellular domain of VAMP2 on the cell surface (CHO-59C) or epitope mutant control cells (CHO-59C-mut) (A); the HBV-infected HepG2-hNTCP cells or non-infected cells (B). The ratio of effector cells to target cells (E:T) was 6:1. The ADCC activity was measured using a lactate dehydrogenase (LDH) release assay, showing as percentages of cytotoxicity. (C) 2H5-A14 induced ADCC and ADCP via macrophages. The target cells used were suspension 293F cells expressing the 2H5-A14 epitope or the mutant epitope, as described above for the CHO cells (A). The effector cells were human blood monocyte-derived macrophages. The E:T ratio was about 1:1. The ADCC activity (left) was measured as described above. For the ADCP assay (right), the target cells were labeled with CFSE fluorescent dye prior to mixing with macrophages. ADCP activity was monitored by fluorescence microscopy. The phagocytosis index was determined as the number of CFSE-positive target cells per 100 macrophages. The means and error bars shown were from two independent experiments. (D) CDC activity of 2H5-A14 and other control antibodies. Rabbit complement sera (10%) (Sigma-Aldrich) were used. All the samples were tested in triplicates. Rituximab (anti-CD20) and GC33 (anti-GPC3) mAbs were positive control mAbs for the assay. They have the same Fc region of human IgG1 as 2H5-A14. All data shown in panel A, B and D represent at least two independent experiments. All the antibodies were tested at 10 μg/mL in panel C-D.
-
Figure 4—source data 1
Data for Figure 4A–D.
Analysis of 2H5-A14 mediated ADCC, ADCP and CDC.
- https://doi.org/10.7554/eLife.26738.025
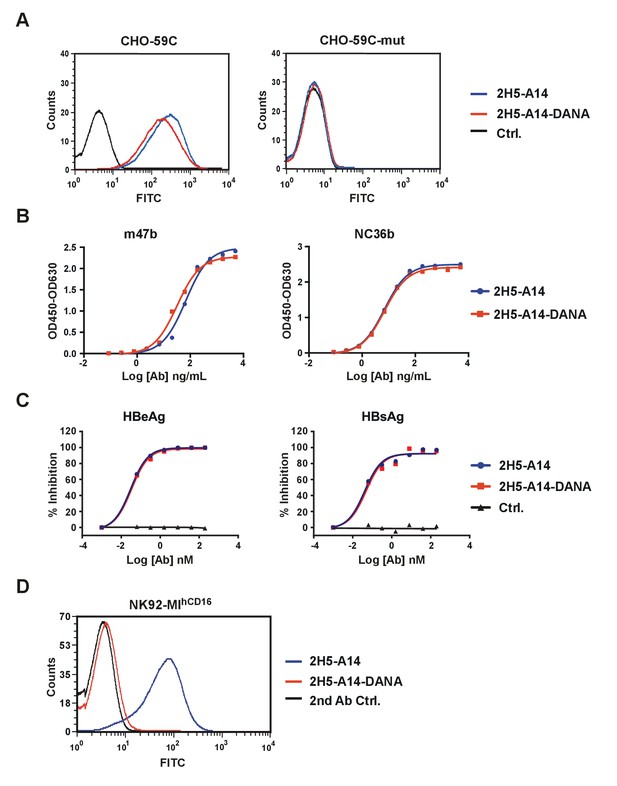
Characterization of 2H5-A14 and its Fc mutant (2H5-A14-DANA) by FACS.
(A) FACS analysis of Ab binding to CHO stable cell lines. CHO-59C cells express the epitope of 2H5-A14 on the cell surface, whereas the CHO-59C-mut cells express a binding-deficient 2H5-A14 epitope mutant (D20A) on their surface. The antibodies (1.1 µg/mL) were incubated with cells, followed by FITC-anti-human Fc antibody staining. 2H5-A14-DANA (D265A/N297A) is a 2H5-A14 mutant of the Fc region that binds to target cells similarly to wild type 2H5-A14. (B) Similar binding activity of 2H5-A14 and 2H5-A14-DANA to preS1 peptides (7.8 nM). (C) Similar neutralization activity of 2H5-A14 and 2H5-A14-DANA against HBV infection of HepG2-hNTCP cells. (D) 2H5-A14-DANA lost binding activity with FcγRIIIa. NK92-MIhCD16 cells expressing exogenous human FcγRIIIa (CD16). The antibodies were tested at 10 µg/mL.
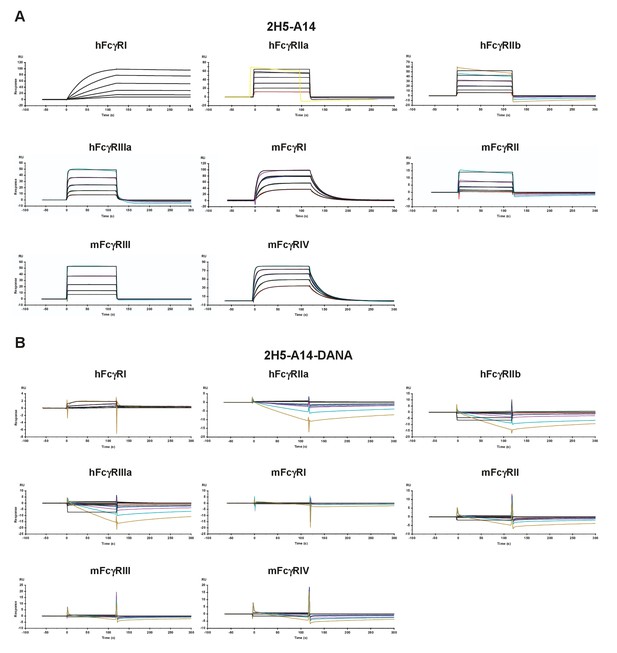
Characterization of the binding of 2H5-A14 and its Fc mutant (2H5-A14-DANA) with human or mouse FcγRs via Biacore analysis.
(A) Binding of 2H5-A14 to human (h) and mouse (m) FcγRs. (B) Binding of 2H5-A14-DANA mutant to human and mouse FcγRs. 2H5-A14-DANA lost the ability to bind with all of the FcγRs tested. 2H5-A14 or 2H5-A14-DANA antibodies were captured on a CM5 chip via Protein A/G that had been covalently immobilized on the chip. FcγRs at various optimized concentrations were injected over the chip surface. Binding kinetics was evaluated using a 1:1 Langmuir binding model.
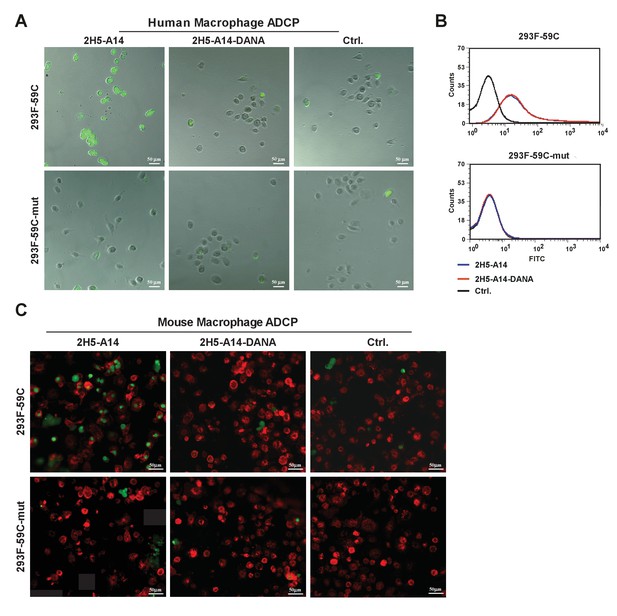
2H5-A14-mediated ADCP via human and mouse macrophages.
(A) ADCP of 293 F-59C cells via human macrophages. 293 F-59C cells express the epitope of 2H5-A14 on the cell surface, whereas the 293F-59C-mut cells express a binding-deficient 2H5-A14 epitope mutant (D20A) on their surface. 293 F-59C and 293F-59C-mut cells were labeled with CFSE fluorescent dye prior to mixing with macrophages. The fluorescence microscopy images shown are representative of two independent experiments. The calculated phagocytosis index is presented in Figure 4C-right. (B) FACS analysis of Ab binding to 293 F-59C cells and 293F-59C-mut cells. (C) ADCP of 293 F-59C cells via mouse macrophages. The target cells, 293 F-59C or 293F-59C-mut, were CFSE-labeled as in panel A, and mouse macrophages were labeled with F4/80-Alex Fluor 647 prior to mixing with target cells. All the antibodies were tested at 10 μg/mL.

CDC and complement C1q binding activity of 2H5-A14 and other antibodies.
(A) CDC. 2H5-A14 did not induce CDC against target cells in the presence of human complement sera (10%) (Sigma). All the samples were tested in triplicates and the antibodies were tested at 10 μg/mL. (B) C1q binding by ELISA. Left, binding with purified human C1q. Human C1q protein (Abcam) were 3-fold serial diluted and tested for binding to different antibodies (10 μg/mL) coated on a 96-well ELISA plate. HRP-labeled anti-human C1q (1:400, Abcam) was used to detect the interaction between testing antibodies and human C1q. Right, binding with mouse serum C1q. FRG mouse sera (10-fold serial diluted) or PBS were added to a 96-well ELISA plate coated with different antibodies (10 μg/mL), followed by rat-anti-mouse C1q Ab (1:500, Abcam) and then by HRP-labeled goat-anti-rat Ab (1:5000, Sigma). OD values of PBS blank controls were subtracted from each of the other readings.
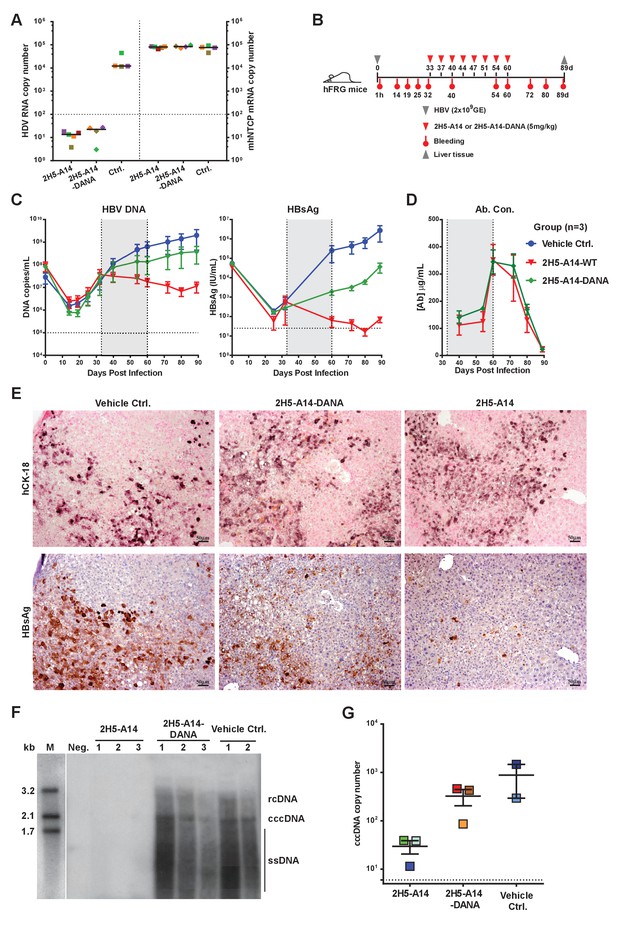
Contributions of Fc-mediated effector functions to the therapeutic effect of 2H5-A14 against HDV and HBV infection in mouse models.
(A) Fc-mediated effector functions are not required for 2H5-A14 to protect mice from HDV infection. The experiment was performed similarly to that described in Figure 3A. Both 2H5-A14 and Fc mutant 2H5-A14-DANA were tested at 5 mg/kg. The copy numbers shown in the Y-axes are from 20 ng of total liver RNA for each sample. (B) Schematic diagram illustrating HBV challenge, bleeding and antibody treatment schedules in the mouse study. Three animals were used in each group. Recipient hFRG mice were challenged with HBV viruses (genotype D) at 2 × 109 GE. Twice weekly treatment with PBS, 2H5-A14 (5 mg/kg), or 2H5-A14-DANA (5 mg/kg) began at 33 dpi and lasted for 4 weeks. (C) HBV DNA titers and HBsAg levels in serum. Blood samples were collected at the indicated time points for measuring HBV DNA titers and/or HBsAg levels. (D) Antibody concentrations. The antibody concentrations in serum samples were measured by ELISA using antibody standards with known concentrations. Note, for dpi 40 and dpi 54, the blood samples were collected at three days after Ab administration; for dpi 60, the blood samples were collected 1 hr after Ab administration. The horizontal dotted lines indicate the lowest detection limits; the vertical dotted lines and the grey-shaded areas indicate the treatment window (C–D). (E) IHC staining of human cytokeratin-18 (hCK18) and HBsAg in serial sections of liver tissues from sacrificed hFRG mice at the end of the experiment (dpi 89). Intrahepatic HBsAg was detected by a specific anti-HBsAg mouse mAb, followed by HRP-anti-mouse secondary Ab and stained brown using DAB substrate, nuclei were stained blue by Hematoxylin. Human hepatocytes in consecutive tissue sections were visualized by staining with a human-specific hCK18 mAb and DAB substrate in blue violet, nuclei were stained with Nuclear Fast Red. Each image shown represents the staining result for one mouse of each group. (F) Southern blot analysis of intrahepatic viral DNA. Total HBV DNA was extracted from mouse liver tissues at dpi 89. The extracted DNA samples were analyzed by Southern blotting with an [α-32P]dCTP-labeled full-length HBV DNA probe. DNA samples prepared from normal mouse livers were used as a negative control. 100 pg each of 3.2 kb, 2.1 kb, and 1.7 kb HBV DNA fragments were used as DNA markers. rcDNA (relaxed circular DNA), cccDNA, and ssDNA (single-strand DNA) intermediates are labeled. (G) qPCR quantification of intrahepatic viral cccDNA level. 500 ng of total DNA prepared from the liver tissues collected at dpi 89 was digested by PSAD and 1/10 of the digested samples were used to quantify HBV cccDNA by qPCR using specific primers (see Materials and methods). The hNTCP gene copy numbers, which represent the amount of human hepatocytes in the liver tissues of chimeric mice, were used to normalize the cccDNA level in the human hepatocytes for each sample. The cccDNA copy number shown in the Y-axis is the relative value normalized to 1000 copies of hNTCP gene in ~50 ng total DNA samples. Each square represents one mouse. The horizontal dotted line indicates the reliable detection limit.
-
Figure 5—source data 1
Data for Figure 5A, 5C-D, and G.
Testing of Fc-mediated effector functions of 2H5-A14 against HDV and HBV infection in mouse models.
- https://doi.org/10.7554/eLife.26738.029
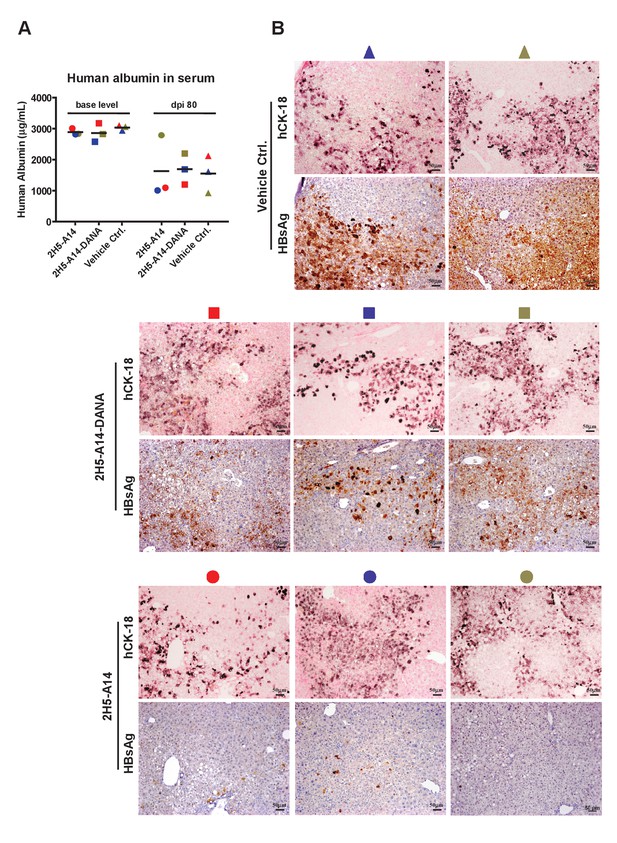
Human albumin levels in hFRG mouse serum and IHC of HBsAg in liver tissues of HBV-infected hFRG mice.
The mice were infected and treated as described in Figure 5. (A) Human albumin levels. Symbols of the same shape and color represent data from the same mouse. The base line levels of human albumin in serum for each mouse were measured at Yecuris after human-liver transplantation. The human albumin levels at dpi 80 for all mice were quantified using a Human Serum Albumin ELISA Kit (Thermo Fisher). (B) IHC staining of hCK18 and HBsAg in serial sections of liver tissues from sacrificed hFRG mice at the end of the experiment. Each set of images shown here represents the staining results of one mouse in each group. A representative set of images for each group is shown in Figure 5E, and is included in this figure as well to enable easier comparisons. Only two sets of images from two mice are shown for the control group, as one mouse in this group was found dead prior to the scheduled euthanasia; no liver tissue was collected for this mouse. Each set of images is also labeled with the same symbol as shown in panel A, indicating the same mouse.

Phylogenetic trees of the protein sequences of the L gene cloned from the liver tissues of HBV-infected hFRG mice.
The sequences were obtained from vehicle control mice (PBS) and 2H5-A14 treated mice, respectively. (A) The mice were infected and treated as described in Figure 3. Liver tissues from two mice of each group were used to clone the L genes. 19 sequences from the PBS group and 22 sequences from the 2H5-A14 treated group were separately obtained and analyzed. (B) The mice were infected and treated as described in Figure 5. 50 sequences from two mice of the PBS group and 63 sequences from the total three mice of the 2H5-A14 treated group were separately obtained and analyzed.
Tables
Data collection and refinement statistics.
Values in parentheses are for the highest-resolution shell.
| preS1-2H5 scFv | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 142.7, 55.7, 67.7 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.9793 |
| Resolution range (Å) | 20–2.5 (2.54–2.50) |
| Unique reflections | 18558(886) |
| Redundancy | 5.8 (5.9) |
| <I > /<σ(I)> | 17.7 (6.2) |
| Completeness (%) | 96.0 (94.9) |
| Rmerge | 0.144 (0.345) |
| Refinement | |
| Resolution range (Å) | 20–2.5 (2.90–2.50) |
| No. reflections | 18265 |
| No. atoms | 3747 |
| Protein | 3519 |
| Water | 227 |
| Ion | 1 |
| Rwork | 0.231 (0.292) |
| Rfree | 0.283 (0.356) |
| Mean B factor (Å2) | 14.5 |
| Rmsd bond length (Å) | 0.002 |
| Rmsd bond angles (°) | 0.555 |
Contact residues between 2H5 and PreS1 peptide.
Contact residues are here defined by interatomic distances of less than 5 Å. Residues involved in salt bridge and hydrogen bond interactions are bolded; the remaining residues engage in van der Waals or hydrophobic interactions.
| 2h5 scFv | HCDR1 | HCDR2 | HCDR3 | CDR-L1 | CDR-L3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A38 | R55 | Y57 | R59 | G107 | Q108 | M109 | G113 | Y37 | Y38 | Y107 | |
| PreS1 | F23 | D20 P21 A22 F23 | A22 | A22 F23 | F23 | F23 | F23 A25 | D20 F23 | P21 N26 | S27 | D20 P21 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.26738.030





