Non-Canonical G-quadruplexes cause the hCEB1 minisatellite instability in Saccharomyces cerevisiae
Figures
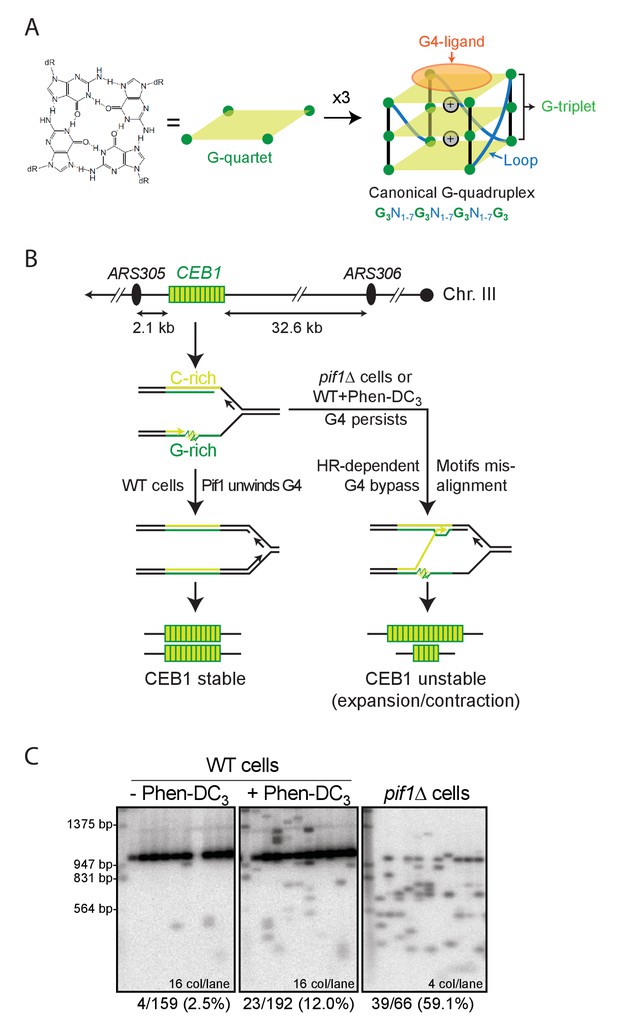
G-quadruplexes and G4-dependent minisatellite instability in S.cerevisiae.
(A) Schematic representation of the overall G4 structure, its features and the underlying canonical G4 motif. N can be any nucleotide. G4-ligands such as Phen-DC3 bind by stacking on an outermost G-quartet. (B) Site of CEB1 integration in the yeast genome, near the replication origin ARS305. CEB1 is oriented so that the G-rich strand is template for the leading strand replication machinery emanating from ARS305. Model for G4-dependent minisatellite instability during leading-strand replication (Lopes et al., 2011). (C) Example of CEB1 instability in untreated or Phen-DC3-treated WT cells and in a pif1Δ mutant. The main band above the 947 bp marker is the parental size CEB1. The Southern blots were published previously in (Piazza et al., 2015).
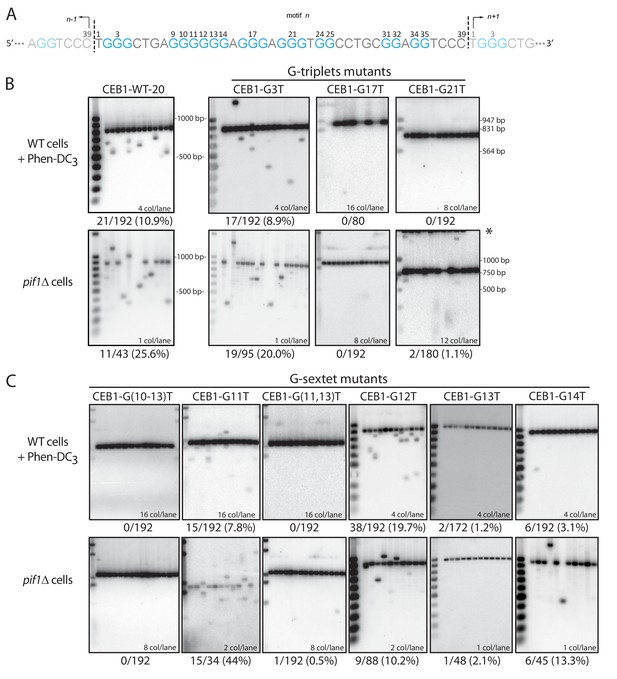
Determination of the G-tract requirements for CEB1 instability in S.cerevisiae.
(A) CEB1 motif with guanine tracts colored in cyan and numbered. (B–C) Southern blot of CEB1-WT-20 and CEB1 variant alleles mutated for their G-tracts in WT cells treated with Phen-DC3 or in pif1Δ cells. (B) G-triplet and (C) G-sextet mutants. In most instances, several independent colonies were pooled and extracted. The number of colonies analyzed per lane and the total rearrangement frequency is indicated for each blot. The fragment sizes (bp) of the molecular ladders run in the first lane of each blot are indicated.
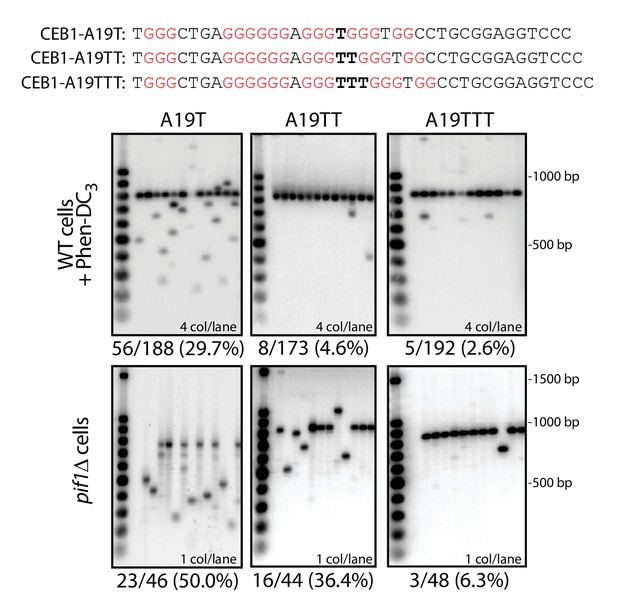
Effect of loop length and sequence on CEB1 instability.
Substitution of the A19 loop located between two essential G-triplets by a Thymine causes an increase in CEB1 rearrangement frequencies both in Phen-DC3-treated WT cells and in pif1Δ cells (ANT1909 and ANT1923, respectively; p-values vs. CEB1-WT-20 < 0.05 in both cases). Increasing the loop length in CEB1-A19TT (strains ANT1910 and ANT1924) and CEB1-A19TTT (ANT1911 and ANT1925) caused a gradual decrease up to non-significant instability levels for CEB1-A19TTT.
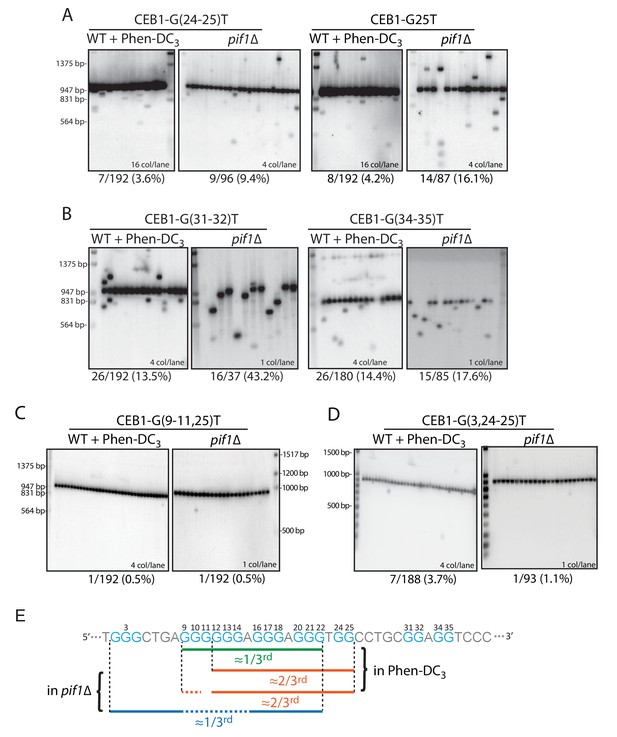
Role of the CEB1 G24-25-doublet in CEB1 instability.
(A) Mutation of the G24-25 doublet in CEB1-G25T or CEB1-G(24-25)T reduced by 2/3rd the minisatellite instability in both pif1Δ (ORT7172 and ORT7173) and Phen-DC3-treated WT cells (ORT7164 and ORT7165). (B) Mutation of the G31-32 and G34-35 doublets did not affect CEB1 instability in pif1Δ cells (ANT1965 and ANT1967) and Phen-DC3-treated WT cells (ANT1950 and ANT1952). (C) Mutation of the G-sextet in CEB1-G(9-11)T-G25T abolished the G24-25-independent instability in both WT Phen-DC3-treated (ANT1973) and pif1Δ (ANT2604) cells. (D) Mutation of the dispensable G2-4 triplet in CEB1-G(3,24–25)T did not decrease the G24-25-doublet-independent instability in Phen-DC3-treated WT cells (ANT2620) but abolished it in a pif1Δ strain (ANT2627). (E) Mapping of the overlapping sets of G-tracts required for full CEB1 instability in the WT Phen-DC3-treated or pif1Δ cells.
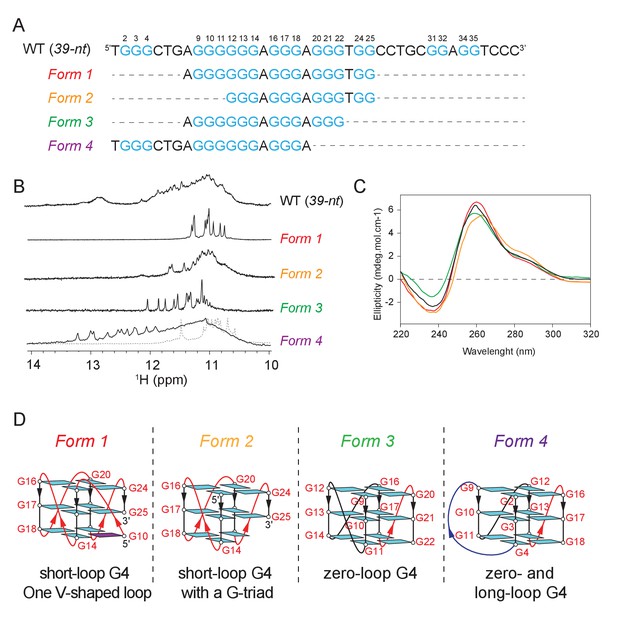
The CEB1 motif adopts multiple non-canonical G4 conformations.
(A) Oligonucleotide sequences used to isolate individual G4 conformation in CEB1. Guanine tracts are colored cyan and numbered within a repeat unit according to their positions. (B) NMR spectra and (C) CD spectra of the mixture of G4 resulted from the CEB1-WT motif and of isolated G4 folds Form 1, 2, 3 and 4. The Form 4 spectra showed in dotted line corresponds to the dimeric form observed at high K+ and DNA strand concentration. The resonance frequency of the nuclei is expressed in part per million (ppM). (D) Isolated G4 folding topologies result from the CEB1 sequence. The snapback guanine in Form 1 can be either G10 or G11, with an associated V-shaped loop containing one or zero G, respectively (Adrian et al., 2014). Form 2 resembles Form 1 but lacks the snapback guanine, and consequently exhibits a terminal G-triad. Form 3 contains a 0-nt loop between the G9-11 and the G12-14 strands. The monomeric Form 4 exhibits a 4-nt long loop between the G2-4 and G9-11 strands, and a 0-nt loop between the G9-11 and G12-14 strands. For the dimeric Form 4 see Figure 4—figure supplement 4J.
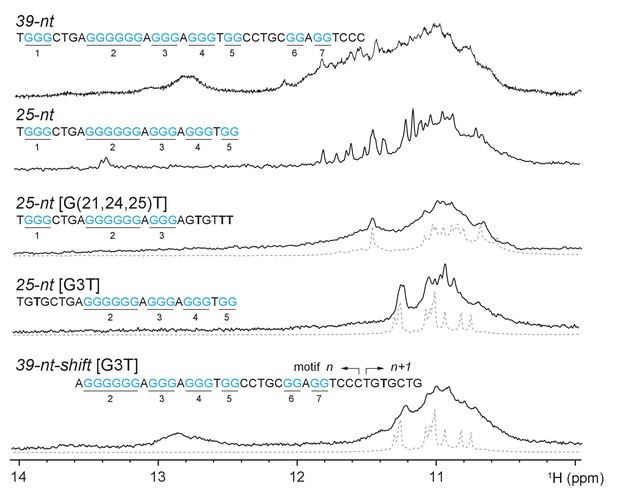
Multiple G4s resulted from different G-rich fragments of CEB1 minisatellite.
Conformational multiplicities were eliminated by residue specific mutations targeted at particular G-tracts. 7-nt truncation from 5’-end of 25-nt [G3T] resulted in Form 1. 6-nt truncation from 3’-end of 25-nt [G(21,24,25)T] resulted in Form 4. Imino proton spectra of Form 1 and Form 4 are shown in gray dotted line below 25-nt [G3T] / Form 3 [G3T] and 25-nt [G(21,24,25)T], respectively. Guanine tracts are colored cyan and numbered according to their positions in 25-nt sequence. G-to-T mutations are highlighted in bold.
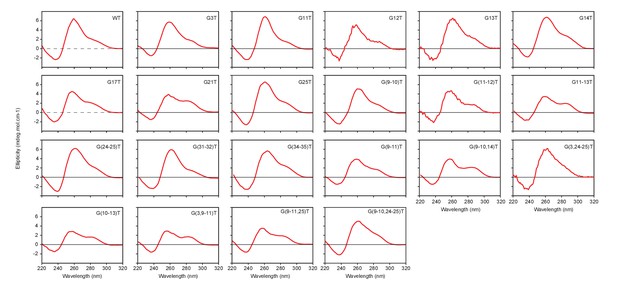
CD spectra of the mutant CEB1 G4 motifs used in this study.
https://doi.org/10.7554/eLife.26884.010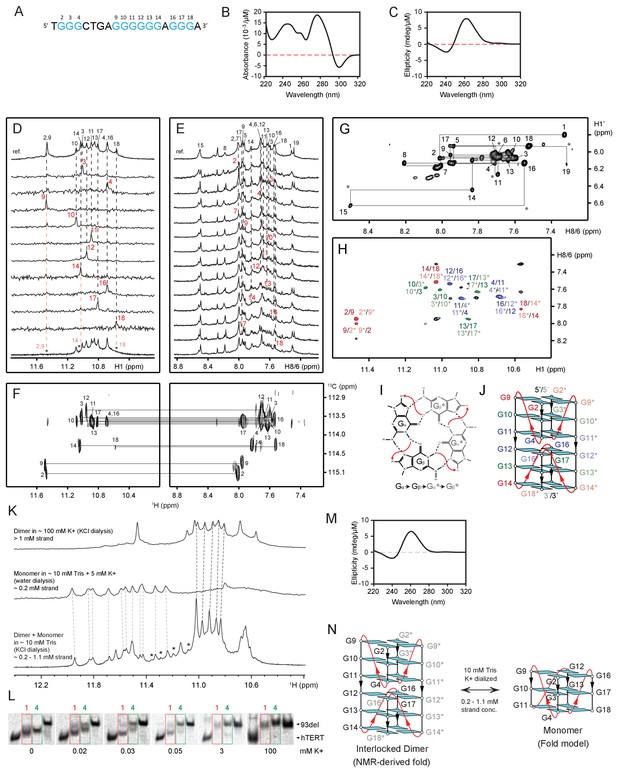
NMR structural characterization of CEB1 Form 4 in 100 mM K+ solution.
(A) The G-rich sequence. (B) TDS and (C) CD spectra. (D) Imino proton assignments from 15N-filtered spectra of samples, 2% 15N-enriched at indicated positions. (D’) Imino proton spectrum following 15 min exposure in D2O solvent at 25°C. Disappearing imino peaks are marked with asterisks. (E) H8 proton assignments by site-specific 2H labeling at the indicated positions. The reference spectra (ref.) of imino and aromatic protons are shown at the top of the corresponding assignment spectra. (F) Through-bond correlations between guanine imino and H8 protons via 13C5 at natural abundance, using long-range J-couplings shown in the inset. (G) The H8/H6-H1’ sequential connectivity on NOESY spectrum (mixing time, 300 ms). (H) The imino-H8 cyclic connectivities on NOESY spectrum (mixing time, 300 ms). The tetrad arrangements (I) were identified from framed cross-peaks with associated label of the residue number of imino proton in the first position and that of H8 proton in the second position. Intra-residue H8/6-H1’ cross-peaks are labeled with residue numbers. Missing cross peaks are marked with asterisks. (J) Schematic representation of interlocked dimeric Form 4. Anti guanines are colored cyan. The backbones of the core and loops are in black and red, respectively. Labels from different strands are in respective dark/light colors. The coexistence of monomeric and dimeric folds of Form 4 was shown using NMR and gel shift assay. (K) NMR spectra of Form 4 under different parameters that is, cation concentration, buffer condition and sample preparation. Imino peaks from unidentified minor form are marked with asterisks. (L) Mobility shift of Form 4 (highlighted in green boxes) in the presence of different concentrations of counter ions visualized by non-denaturing PAGE. Mobility shifts of Form 1 are highlighted in red boxes. (M) CD spectrum of monomeric Form 4 recorded in 5 mM K+ solution at ~4 μM strand concentration. (N) Schematic representation of NMR-derived interlocked dimeric and model monomeric Form 4.
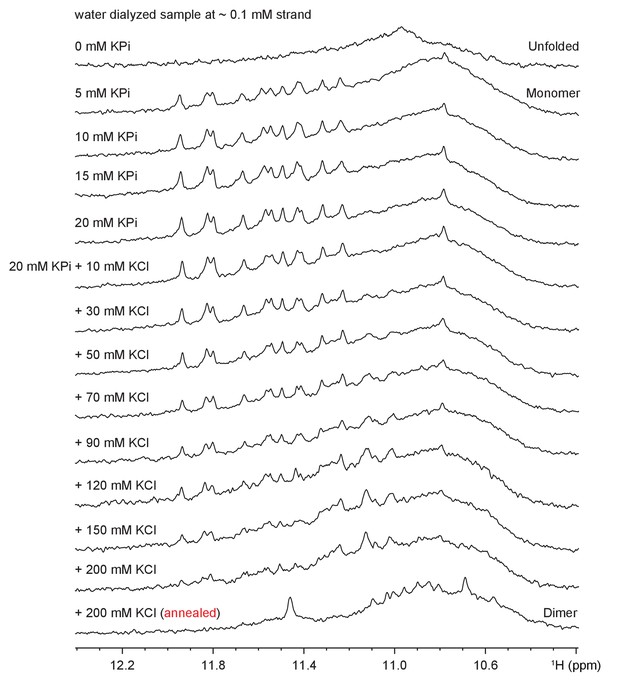
Imino spectral transition of Form 4 through K+ titration at ~0.1 mM strand concentration.
Full transition to dimeric G4 was reached following sample annealing at >200 mM K+.
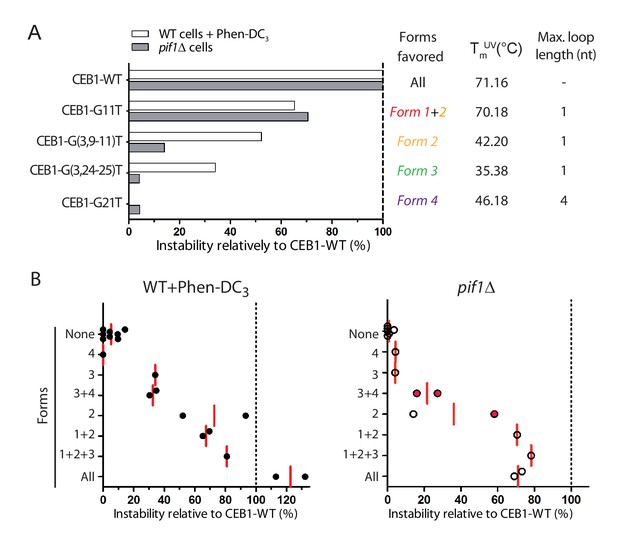
Contribution of G4 conformations to CEB1 instability.
(A) Example of instabilities obtained in Phen-DC3-treated WT cells and in pif1Δ cells for CEB1 variants isolating single G4 conformations, normalized to the instability obtained for CEB1-WT. Since the sequence requirements for Form 2 is embedded in the sequence requirements for Form 1, no mutation could isolate Form 1. Its contribution can be deduced by comparing with alleles isolating Form 2. Thermal stability and maximum loop length for each isolated form are indicated. (B) Summary of the CEB1 variants instability in Phen-DC3-treated WT cells and in pif1Δ cells as a function of the forms they can adopt. Each point corresponds to a different allele. The mean is shown in red. Red dots indicate possible formation of an unidentified form involving G2-4.
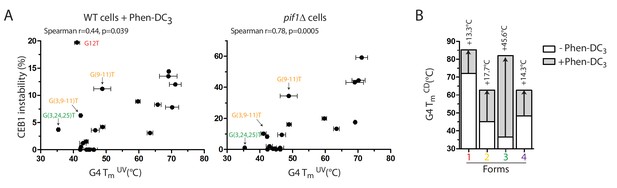
Correlation of CEB1 instability and G4 thermal stability.
(A) CEB1 variant instabilities correlate with the thermal stabilities of their associated G4s as determined by UV-melting, in both WT cells treated with Phen-DC3 (left) and in pif1Δ cells (right). (B) Phen-DC3 differently stabilizes the isolated G4s resulting from CEB1, as determined by CD-melting. Arrows indicate the ΔTm.
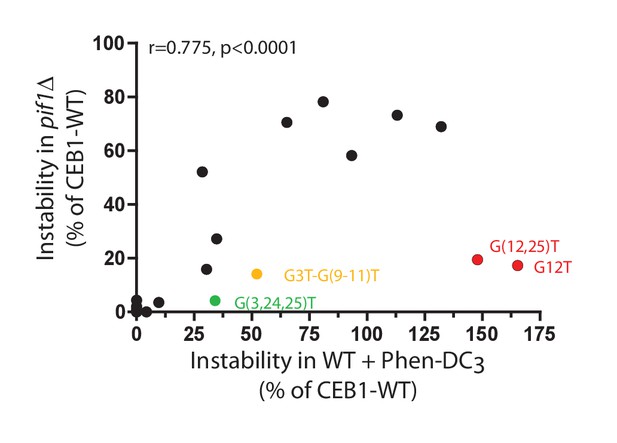
Correlation between CEB1 variant instabilities upon Phen-DC3 treatment and PIF1 deletion.
Genomic instabilities measured in pif1Δ strains and in Phen-DC3-treated WT strains are significantly correlated. Notable deviations are highlighted. Instabilities are represented as a percentage of CEB1-WT instability.
Tables
Genomic instabilities of CEB1 variants and the associated G4 thermal stability.
| Allele | Motifs | Sequence | WT cells | WT cells + Phen-DC3 | pif1D cells | Forms | G4 TmUV (°C) |
|---|---|---|---|---|---|---|---|
| CEB1-1.8 (Lopes et al., 2011) | 42 | 0.5% (384) | 11.2% (992) | 56.3% (119) | |||
| WT-25 (Piazza et al., 2015) | 25 | TCTGAAATCCTGCATCCC | 2.5% (159) | 12.0% (192)* | 59.1% (66)* | all | 71.16 (1.78) |
| WT-20 | 20 | 0 (96) | 10.9% (192)* | 25.6% (43)* | |||
| WT-10 | 10 | 0 (96) | 4.2% (176) | ND | |||
| Single G-tracts mutations | |||||||
| Gmut (Ribeyre et al., 2009) | 20 | TCCTGACCATAATCCTGCGGAGGTCCC | 1.0% (96) | 1.0% (192) | ND | - | NA |
| 42 | TCCTGACCATAATCCTGCGGAGGTCCC | ND | ND | 0 (384) | |||
| G3T | 22 | TTCTGAAATCCTGCGGAGGTCCC | 1.0% (96) | 8.9% (192)* | 20.0% (95)* | 1, 2, 3 | 59.74 (0.74) |
| G(9,10)T | 24 | TCTGATTAATCCTGCGGAGGTCCC | 0 (92) | 8.3% (192)* | ND | 1, 2 | 65.8 (0.95) |
| G(9-11)T | 24 | TCTGATTTAATCCTGCGGAGGTCCC | 0 (96) | 11.2% (188)* | 34.4% (64)*† | 2 | 48.81 (2.65) |
| G(9,10,14)T | 21 | TCTGATTTAATCCTGCGGAGGTCCC | 0 (192) | 0 (192)† | 0.5% (192)† | (2) | 44.74 (0.31) |
| G(10-13)T | 25 | TCTGATTTTAATCCTGCGGAGGTCCC | 0 (176) | 0 (192)† | 0 (192)† | - | 46.24 (1.08) |
| G11T | 25 | TCTGATAATCCTGCGGAGGTCCC | 0.5% (192) | 7.8% (192)* | 44.4% (36)* | 1, 2 | 70.18 (2.00) |
| G(11-12)T | 24 | TCTGATTAATCCTGCGGAGGTCCC | 0 (96) | 0.5% (192) | ND | - | ND |
| G(11,13)T | 26 | TCTGATTAATCCTGCGGAGGTCCC | 0 (192) | 0 (192)† | 0.5% (192)† | - | 44.02 (0.78) |
| G12T | 24 | TCTGATAATCCTGCGGAGGTCCC | 0 (92) | 19.7% (192)*† | 10.2% (88)*† | - | 41.11 (0.81) |
| G13T | 24 | TCTGATAATCCTGCGGAGGTCCC | 0 (96) | 1.2% (172)† | 2.1% (48)† | - | 42.92 (0.98) |
| G14T | 20 | TCTGATAATCCTGCGGAGGTCCC | 0 (96) | 3.1% (192)† | 13.3% (45)*† | (1, 2) | 63.44 (0.92) |
| G17T | 24 | TCTGAATATCCTGCGGAGGTCCC | 0 (128) | 0 (80)† | 0 (192)† | - | 44.97 (0.52) |
| G21T | 19 | TCTGAAATTCCTGCGGAGGTCCC | 0.5% (180) | 0 (192)† | 1.0% (180)† | 4 | 46.18 (1.14) |
| G(24-25)T | 26 | TCTGAAATTTCCTGCGGAGGTCCC | 0 (192) | 3.6% (192)*† | 9.4% (96)*† | 3, 4 | 46.68 (1.21) |
| G25T | 26 | TCTGAAATGTCCTGCGGAGGTCCC | 0 (192) | 4.2% (192)*† | 16.1% (87)*† | 3, 4 | 48.91 (0.71) |
| G(31-32)T | 24 | TCTGAAATCCTGCTTAGGTCCC | 1.0% (96) | 13.5% (192)* | 43.2% (37)* | All | 69.03 (2.68) |
| G(34-35)T | 21 | TCTGAAATCCTGCGGATTTCCC | 5.0% (96) | 14.4% (180)* | 17.6% (85)* | All | 69.16 (0.22) |
| Double G-tracts mutations | |||||||
| G(3,9-11)T | 24 | TTCTGATTTAATCCTGCGGAGGTCCC | 0 (92) | 6.3% (192)* | 8.3% (48)† | 2 | 42.20 (0.33) |
| G(3,24-25)T | 22 | TTCTGAAATTTCCTGCGGAGGTCCC | 0 (96) | 3.7% (188)† | 1.1% (93)† | 3 | 35.38 (0.29) |
| G(9-10,24-25)T | 20 | TCTGATTAATTTCCTGCGGAGGTCCC | 0 (96) | 1.5% (192)† | ND | - | 43.92 (0.65) |
| G(9-11,25)T | 24 | TCTGATTTAATGTCCTGCGGAGGTCCC | 0 (96) | 0.5% (192)† | 0 (48)† | - | 42.32 (0.37) |
| G(12,25)T | 24 | TCTGATAATTCCTGCGGAGGTCCC | 0 (96) | 17.7% (192)* | 11.5% (96)*† | - | ND |
| G(14,24-25)T | 20 | TCTGATAATTTCCTGCGGAGGTCCC | 0 (96) | 0 (188)† | ND | - | 42.10 (0.49) |
-
* p-values vs. untreated WT < 0.05.
-
† p-value vs. CEB1-WT < 0.05.
-
Forms in parenthesis denote a loop modification.
-
Numbers in parenthesis indicate total number of colonies tested.
-
In the Tm column, the number in parenthesis indicates the standard deviation of the triplicate Tm measurement.
-
ND: Not determined; NA: Not applicable
Additional files
-
Supplementary file 1
Strains and primers used in this study.
(A) Relevant genotypes of haploid W303 Saccharomyces cerevisiae strains used in this study. (B) List of CEB1 sequences analyzed using NMR spectroscopy in vitro.
- https://doi.org/10.7554/eLife.26884.016





