Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts
Figures
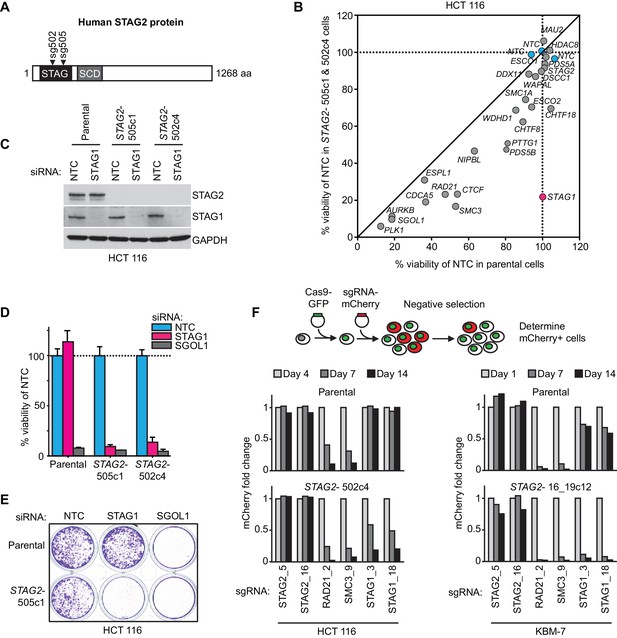
Identification of STAG1 as a genetic vulnerability of STAG2 mutated cells.
(A) Engineering of an isogenic HCT 116 cell model by CRISPR/Cas9-mediated inactivation of STAG2. The position of the sgRNAs used to create deleterious insertion and deletion mutations in the STAG2 coding sequence is indicated. (B) Parental HCT 116 cells and two STAG2 mutant clones (STAG2-) were subjected to an siRNA screen. Pools of 4 siRNA duplexes targeting 25 known cohesin subunits and regulators were transfected into the three cell lines. Cell viability was measured 7 days after transfection. Following normalization to non-target control (NTC) values, the average cell viability of siRNA pools in parental HCT 116 cells and two STAG2- clones was plotted against each other (n = 2 or more independent experiments with 2 biological repeats each). (C) HCT 116 parental cells and STAG2- clones were transfected with the indicated siRNAs. Protein lysates were prepared 72 hr after transfection and analyzed by immunoblotting. (D) Cell viability was assessed 8 days after siRNA transfection using a metabolic assay (n = 4 biological repeats, error bars denote standard deviation) and (E) using crystal violet staining. (F) Cas9-GFP expressing isogenic parental and STAG2- HCT 116 and KBM-7 cells were transduced with a lentivirus encoding mCherry and sgRNAs targeting the indicated genes. The percentage of mCherry-positive cells was determined over time by flow cytometry and normalized to the fraction of mCherry-positive cells at the first measurement and sequentially to a control sgRNA (n = 1 experimental replicate).
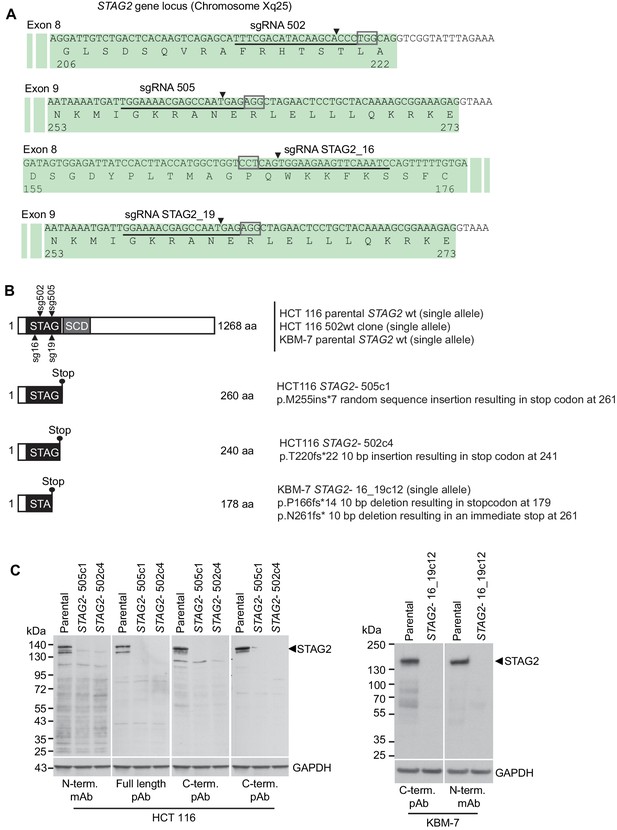
Characterization of CRISPR/Cas9-generated STAG2 mutated clones used in this study.
(A) Four different sgRNAs co-expressed with Cas9 from an all-in-one plasmid or a lentiviral vector were used to generate deleterious frameshift insertions or deletion mutations (indels) in STAG2 coding exons. sgRNAs sg502 and sg16 target exon 8 of STAG2, while sgRNAs sg505 and sg19 target exon 9 of the gene. Cognate sgRNA target sequences are underlined. Associated protospacer adjacent motifs (PAMs) are marked by boxes. The predicted Cas9 cleavage sites are marked by triangles. (B) CRISPR/Cas9-generated STAG2 indels in HCT 116 and KBM-7 cell clones as detected by Sanger sequencing. STAG2 is located on the X chromosome. Therefore, only one allele needed to be inactivated in HCT 116 (male) and KBM-7 (near-haploid) cells. (C) Protein lysates prepared from the indicated parental and mutant clones were analyzed for STAG2 protein expression by immunoblotting using antibodies directed against different STAG2 epitopes.
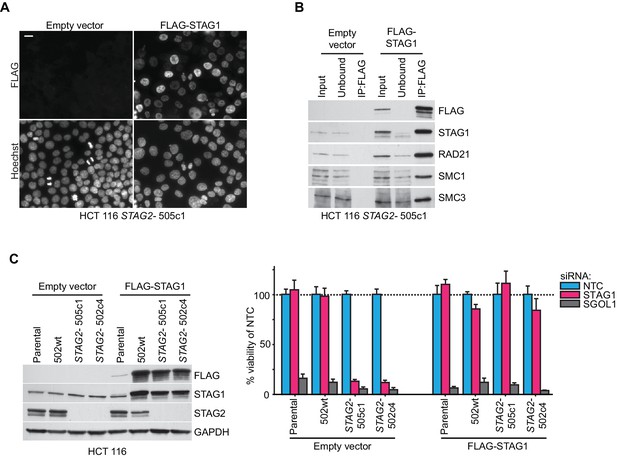
Rescue of the synthetic lethal interaction between STAG1 and STAG2 by expression of an siRNA-resistant FLAG-STAG1 transgene.
HCT 116 parental cells, a STAG2 wild-type clone (502wt), and two STAG2- clones (505c1 and 502c4) were transduced with a lentivirus encoding no transgene (empty vector) or an siRNA-resistant and 3xFLAG-tagged STAG1 transgene (FLAG-STAG1). Stably selected cell pools were used for the analysis. (A) Immunofluorescence analysis of FLAG-STAG1 transgene expression and nuclear localization in HCT 116 STAG2- 505c1 cells. Scale bar, 20 μm. (B) Protein extracts prepared from HCT 116 STAG2- 505c1 cells expressing no transgene (empty vector) or a FLAG-STAG1 transgene were subjected to anti-FLAG immunoprecipitation. The input, unbound and precipitated fractions were analyzed by immunoblotting. Co-precipitation of cohesin subunits was only detected in FLAG-STAG1 expressing cells indicating specific incorporation of the transgenic protein into the cohesin complex. (C) Protein extracts prepared from the indicated cell lines that were transduced with an empty vector or a FLAG-STAG1 transgene were analyzed by immunoblotting (left panel) and transfected with NTC, STAG1 and SGOL1 siRNA duplexes (right panel). Cell viability was measured 7 days after transfection and is plotted normalized to the viability of NTC siRNA transfected cells (n ≥ 5 biological repeats, error bars denote standard deviation).
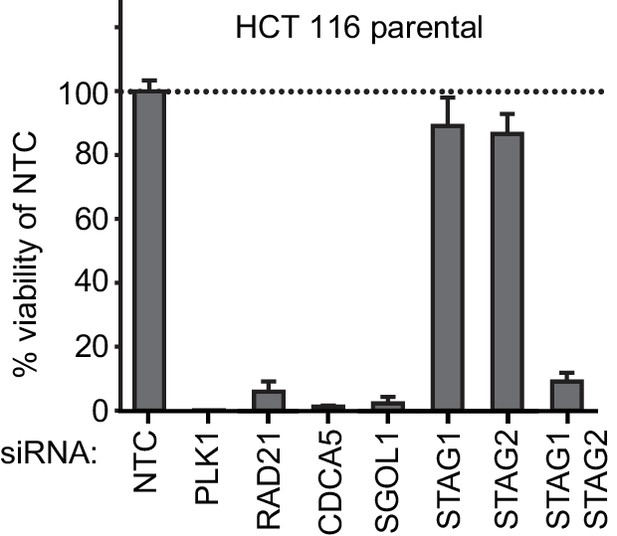
Double depletion experiment confirms STAG1-STAG2 synthetic lethality.
(A) HCT 116 parental cells were co-transfected with NTC siRNA and siRNA duplexes targeting one of the following genes: NTC, PLK1, RAD21, CDCA5, SGOL1, STAG1 or STAG2. HCT 116 parental cells were also co-transfected with siRNA duplexes targeting STAG1 and STAG2. Cell viability was determined 8 days after transfection and is plotted normalized to cell viability of NTC siRNA-transfected cells (n = 3 biological repeats, error bars denote standard deviation).
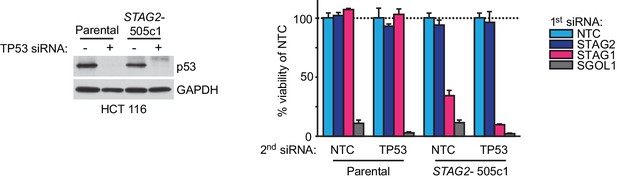
Double depletion experiments indicate that the STAG1-STAG2 genetic interaction is independent of p53.
Parental HCT 116 cells and STAG2- 505c1 cells were transfected with NTC (-) or TP53 (+) siRNA duplexes. Protein extracts were prepared 4 days after transfection and analyzed by immunoblotting (left panel). Parental HCT 116 cells and STAG2- 505c1 cells were co-transfected with the indicated siRNA duplexes (right panel). Cell viability was determined 8 days after transfection and is plotted normalized to the cell viability of NTC+NTC or NTC+TP53 siRNA-co-transfected cells (n = 3 biological repeats, error bars denote standard deviation).
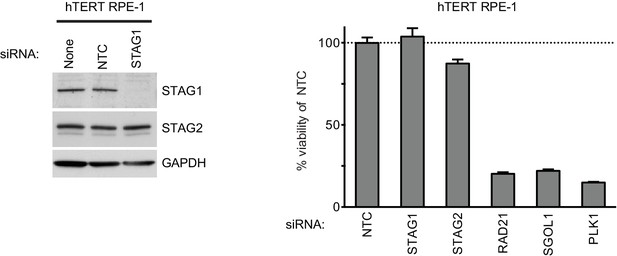
Depletion of STAG1 does not reduce viability in hTERT RPE-1 cells.
Human telomerase-immortalized retinal pigment epithelial cells (hTERT RPE-1) were transfected with the indicated siRNA duplexes. Protein extracts were prepared 4 days after transfection and analyzed by immunoblotting (left panel). Cell viability was determined 5 days after transfection and is plotted normalized to the cell viability of NTC siRNA-transfected cells (n = 2 independent experiments with 3 biological replicates each, error bars denote standard deviation).
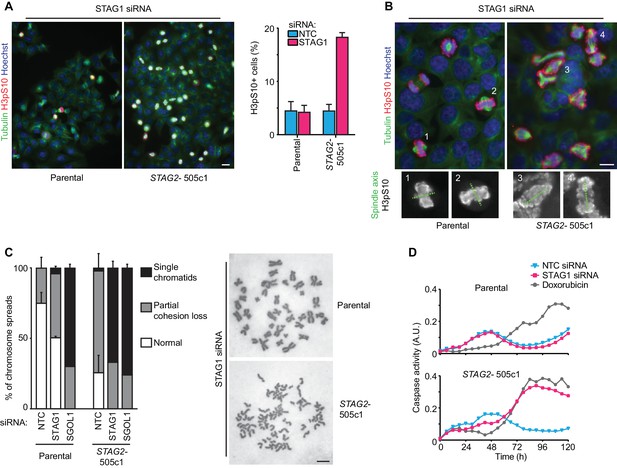
Loss of STAG1 function causes severe mitotic defects, abrogates sister chromatid cohesion and triggers apoptosis in STAG2- but not parental HCT 116 cells.
(A) Parental and STAG2- 505c1 HCT 116 were transfected with NTC and STAG1 siRNA duplexes. Immunofluorescence analysis was performed 72 hr after transfection to determine the mitotic index by scoring the fraction of histone H3 phosphoSer10-positive (H3pS10+) cells (n ≥ 1323 cells, error bars denote standard deviation of three independent experiments), and (B) to investigate mitotic spindle geometry and chromosome alignment. Cropped and magnified examples of chromosome alignment are shown in (B). Scale bars, 20 μm. (C) Giemsa-stained mitotic chromosome spreads were prepared 48 hr after transfection of parental and STAG2- HCT 116 cells with the indicated siRNA duplexes. The status of sister chromatid cohesion of individual metaphase spreads was categorized into normal, partial loss of cohesion or single chromatid phenotypes (n = 100 spreads, error bars denote standard deviation of two independently analyzed slides). Scale bar, 10 μm. (D) Caspase activity was tracked over time using a live-cell caspase 3/7 substrate cleavage assay in parental and STAG2- 505c1 HCT 116 cells after transfection with the indicated siRNAs or after treatment with 0.3 μM doxorubicin at t = 0 hr (n = 2 independent experiments with 4 biological repeats for NTC and STAG1 siRNA each and 1 biological repeat for doxorubicin each).
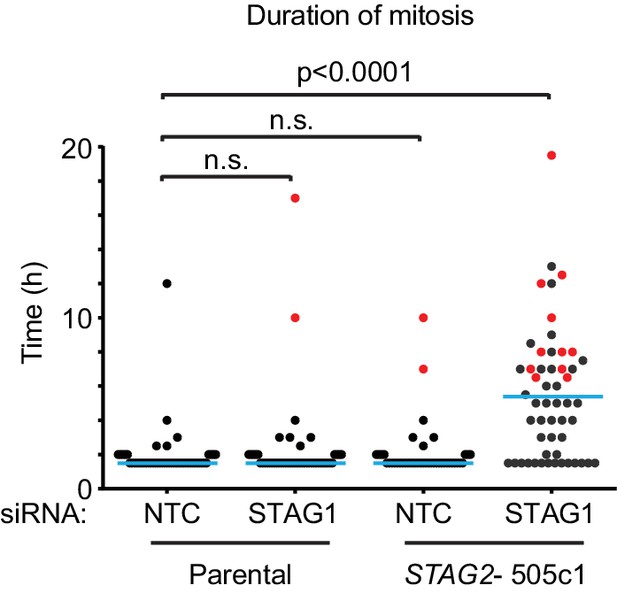
The depletion of STAG1 prolongs mitosis in STAG2 mutated but not parental HCT 116 cells.
Parental and STAG2- 505c1 HCT 116 were transfected with NTC and STAG1 siRNA duplexes. Cells were tracked from 0.5 to 72 hr (imaging interval 30 min) after transfection by bright-field live-cell imaging. The duration of mitosis (time elapsed from mitotic cell rounding to anaphase onset) and the fate of individual cells were determined (n ≥ 43 cells). The duration of mitosis of individual cells is plotted. Blue lines denote mean, red dots denote cells that died in mitosis. Significance levels were quantified using unpaired t test.
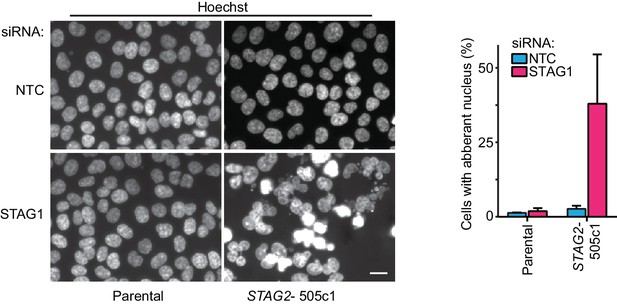
Aberrant nuclear morphology in STAG2-mutated but not parental HCT 116 cells upon depletion of STAG1.
Parental and STAG2- 505c1 HCT 116 were transfected with NTC and STAG1 siRNA duplexes, fixed and stained with Hoechst 72 hr after transfection. Nuclei were scored for aberrant size (>20 μm) or morphology (multi-nucleated cells and cells with polylobed nuclei; n ≥ 612 nuclei, error bars denote standard deviation between two independent experiments).
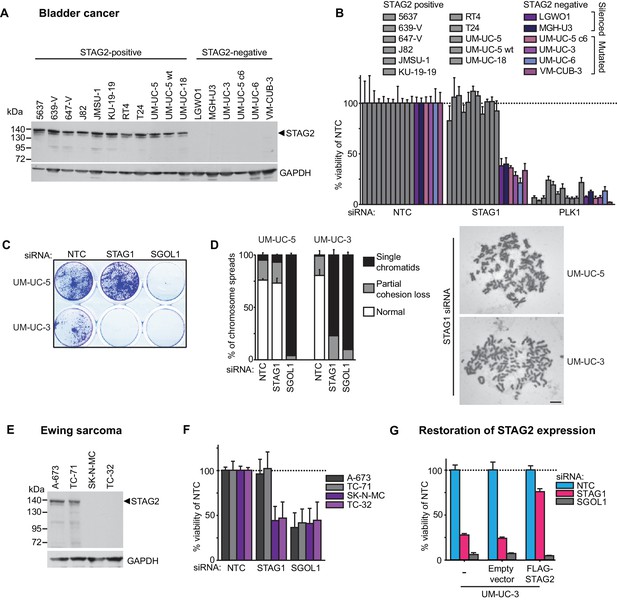
The synthetic lethal interaction between STAG1 and STAG2 is manifested in disease-relevant bladder cancer and Ewing sarcoma cell lines.
(A) The indicated bladder cancer cell lines were analyzed for STAG2 expression by immunoblotting. (B) The indicated bladder cancer cell lines were transfected with NTC, STAG1 and PLK1 siRNA duplexes. Viability was determined 7 or 10 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 2 independent experiments with 5 biological repeats each, error bars denote standard deviation). (C) STAG2 wild-type UM-UC-5 and STAG2 mutated UM-UC-3 cells were transfected with NTC, STAG1 and SGOL1 siRNA duplexes. Colony formation was analyzed 7 days after transfection by crystal violet staining. (D) 72 hr after siRNA transfection into UM-UC-5 and UM-UC-3 cells, Giemsa-stained chromosome spreads were prepared and analyzed for sister chromatid cohesion phenotypes (n = 100 spreads, error bars denote standard deviation of two independently analyzed slides). (E) The indicated Ewing sarcoma cell lines were analyzed for STAG2 protein expression by immunoblotting. (F) The indicated Ewing sarcoma cell lines were transfected with NTC, STAG1 and SGOL1 siRNA duplexes. Viability was measured 6 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 3 independent experiments with 3 biological replicates each, error bars denote standard deviation). (G) STAG2 mutated UM-UC-3 cells were transduced with a lentivirus encoding a FLAG-STAG2 transgene. Stably selected cell pools were subsequently transfected with NTC, STAG1 or SGOL1 siRNA duplexes. Viability was measured 7 days after transfection and normalized to the viability of NTC siRNA transfected cells (n = 4 biological replicates, error bars denote standard deviation).
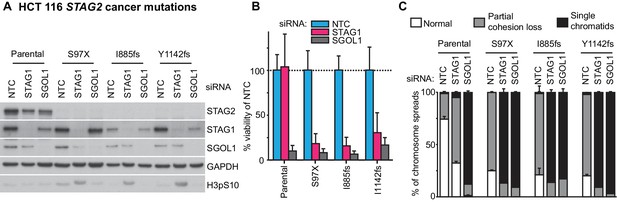
Patient-derived STAG2 mutations cause STAG1 dependency in engineered isogenic HCT 116 cells.
HCT 116 cell lines engineered to harbor the indicated deleterious patient-derived STAG2 mutations were transfected with NTC, STAG1 and SGOL1 siRNA duplexes. (A) Protein extracts were prepared 48 hr after transfection and analyzed by immunoblotting. (B) Cell viability was measured 7 days after siRNA transfection and plotted normalized to the viability of NTC-transfected cells (n = 4 independent experiments with 5 biological repeats each, error bars denote standard deviation). (C) Sister chromatid cohesion phenotypes were analyzed in Giemsa-stained mitotic chromosome spreads that were prepared 48 hr after siRNA transfection (n = 100 chromosome spreads, error bars denote standard deviation between two independently analyzed slides).
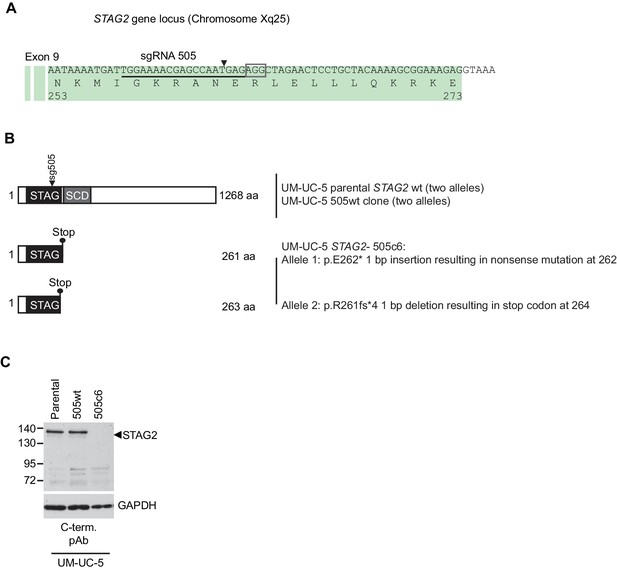
Characterization of CRISPR/Cas9-generated STAG2 knockout in UM-UC-5 bladder cancer cell line.
(A) sgRNA 505 co-expressed with Cas9 from an all-in-one plasmid was used to generate deleterious frameshift insertions or deletion mutations (indels) in STAG2 coding exons. The cognate sgRNA target sequence is underlined. Associated protospacer adjacent motifs (PAMs) are marked by boxes. The predicted Cas9 cleavage site is marked by a triangle. (B) CRISPR/Cas9-generated STAG2 indels in the UM-UC-5 cell clone STAG2- 505c6 as detected by Sanger sequencing. STAG2 is located on the X chromosome. Therefore, two alleles had to be inactivated in UM-UC-5 (female) cells. (C) Protein lysates prepared from the indicated parental and mutant UM-UC-5 clones were analyzed for STAG2 protein expression by immunoblotting.
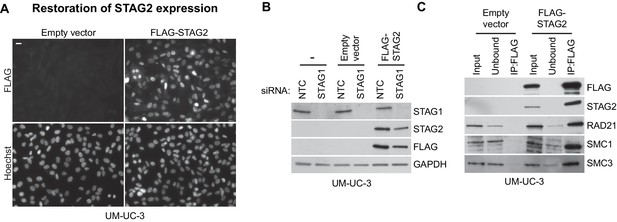
Restoration of STAG2 protein expression in UM-UC-3 bladder cancer cells.
STAG2-deficient UM-UC-3 cells were transduced with lentiviral particles encoding no transgene (empty vector) or a 3xFLAG-tagged STAG2 transgene (FLAG-STAG2). Stable selected cell pools were used for further analysis. (A) Expression and nuclear localization of the transgenic FLAG-STAG2 protein was assayed by immunofluorescence microscopy. (B) Non-transduced UM-UC-3 cells, UM-UC-3 cells transduced with an empty vector and UM-UC-3 cells transduced with a FLAG-STAG2 transgene were transfected with NTC or STAG1 siRNA duplexes. Protein lysates prepared 72 hr after siRNA transfection were analyzed by immunoblotting. (C) Protein extracts prepared from UM-UC-3 cells expressing no transgene (empty vector) or a FLAG-STAG2 transgene were subjected to anti-FLAG immunoprecipitation. The input, unbound and precipitated fractions were analyzed by immunoblotting. Co-precipitation of cohesin subunits was only detected in FLAG-STAG2 expressing cells indicating successful incorporation of the transgenic STAG2 protein into the cohesin complex.
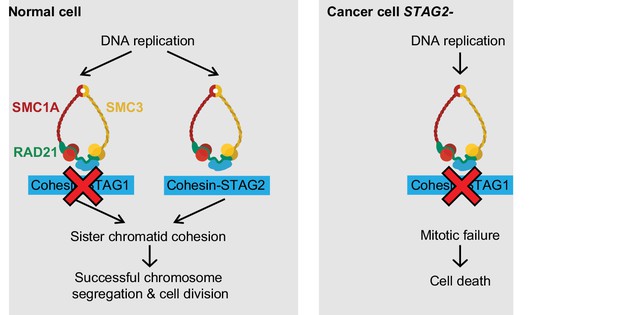
Model for the synthetic lethal interaction between STAG1 and STAG2.
In wild-type cells, both cohesin-STAG1 and cohesin-STAG2 complexes redundantly contribute to sister chromatid cohesion and successful cell division. Loss of STAG1 is tolerated in these cells as cohesin-STAG2 complexes alone suffice to support sister chromatid cohesion for cell division. In cancer cells in which STAG2 is mutationally or transcriptionally inactivated, sister chromatid cohesion is now entirely dependent on cohesin-STAG1 complexes. Inactivation of STAG1 in STAG2 mutated cells therefore results in a loss of sister chromatid cohesion followed by mitotic failure and cell death.
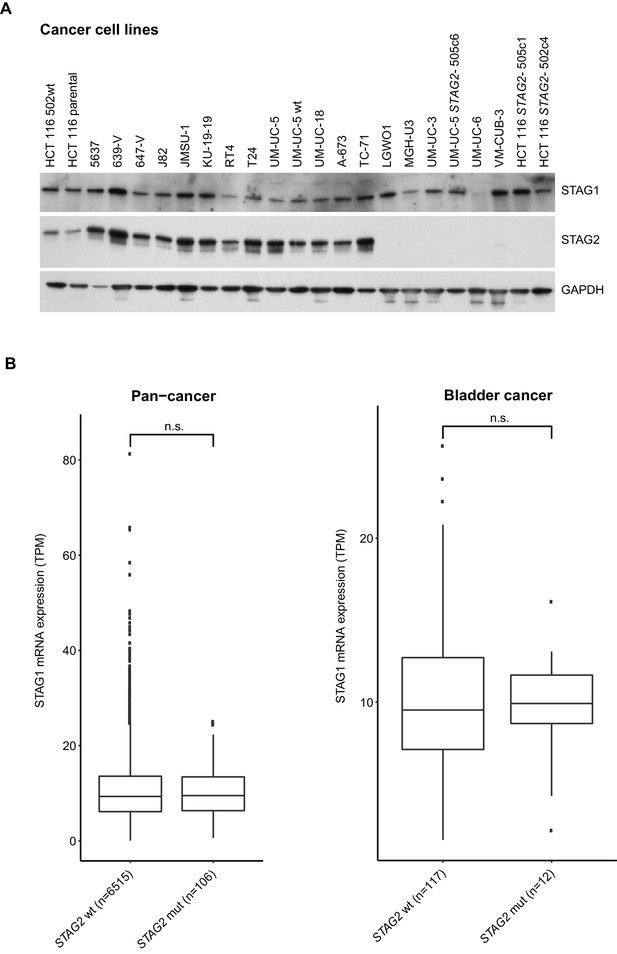
Analysis of STAG1 protein or mRNA expression in cancer cell lines and patient tumors.
(A), Immunoblot analysis of protein extracts prepared from the indicated cancer cell lines. (B), STAG1 mRNA expression levels (transcripts per million, TPM) in tumor samples obtained from The Cancer Genome Atlas (TCGA) pan-cancer datasets (left graph) and bladder cancer datasets (right graph). Genetic alteration data were obtained from cBioPortal. The results shown here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. Samples containing alterations resulting in STAG2 amino acid changes (missense mutations, nonsense point mutations or small insertions/deletions) were catagorized as STAG2 mutated (STAG2 mut). The boxplot bodies indicate the first quartile (25%), median, and third quartile (75%). The whiskers extend no further than 1.5 times the distance between the first and the third quartiles to the lowest and largest extremes from the boxplot bodies. All samples exceeding the whisker limits are shown individually as points. Significance tests were performed with the wilcox.test function of the ggsignif v0.2.0 R v3.3.2 package. Significance levels were labeled as follows: n.s.>0.05.
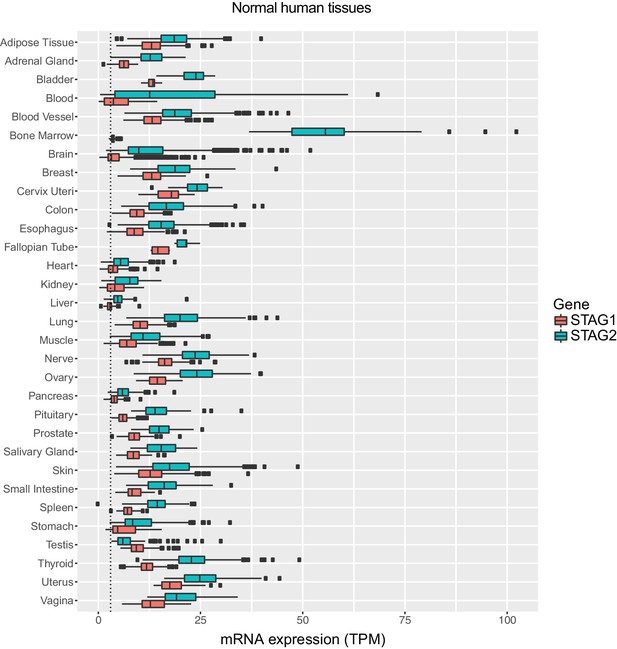
Expression levels of STAG1 and STAG2 mRNA in normal human tissues.
Expression data were obtained from The Genotype-Tissue Expression (GTEx) project (www.gtexportal.org). STAG1 and STAG2 mRNA expression levels (transcripts per million, TPM) in the indicated normal human tissues are plotted. The boxplot bodies indicate the first quartile (25%), median, and third quartile (75%). The whiskers extend no further than 1.5 times the distance between the first and the third quartiles (inter-quartile range, IQR) to the lower and larger extremes from the boxplot bodies. All samples exceeding the whisker limits are shown individually as points. The dotted line indicates a TPM value of 3.
Additional files
-
Supplementary file 1
Table of sgRNA sequences used in this study.
- https://doi.org/10.7554/eLife.26980.019
-
Supplementary file 2
Table of cell lines used in this study.
Sources, STAG2 status and authentication information (STR fingerprinting) of cell lines are listed.
- https://doi.org/10.7554/eLife.26980.020





