Characterisation of the biflavonoid hinokiflavone as a pre-mRNA splicing modulator that inhibits SENP
Figures
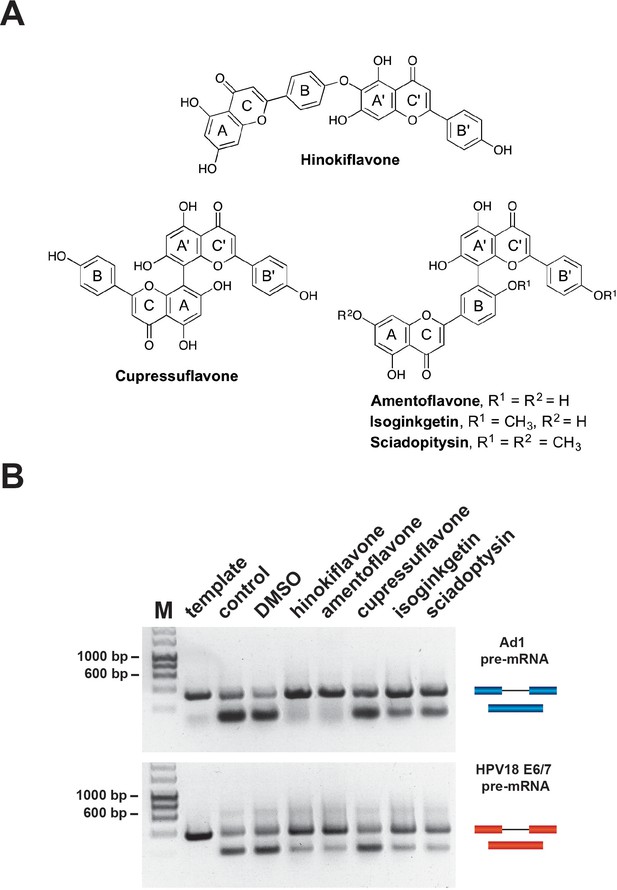
Biflavones inhibit splicing in vitro.
(A) Chemical structures of the five biflavones: hinokiflavone, amentoflavone, cupressuflavone, isoginkgetin and sciadopitysin. These were each tested for their ability to inhibit splicing of the Ad1 and the HPV18 E6/E7 pre-mRNAs in a nonradioactive RT-PCR based in vitro splicing assay. (B) Splicing assays show that all compounds except cupressuflavone inhibited splicing to varying degrees in this assay system.
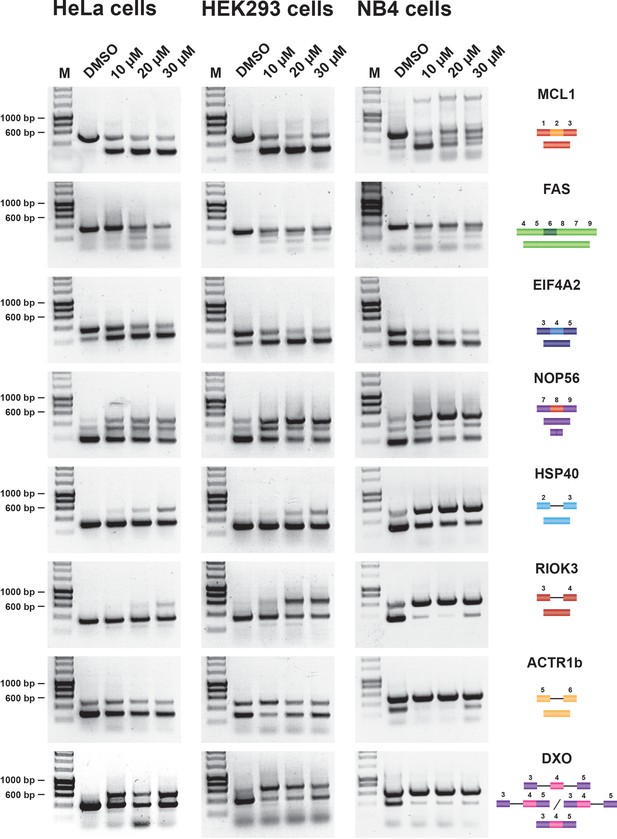
Hinokiflavone modulates splicing in cells.
Semiquantitative RT-PCR analysis of cells treated with increasing concentrations of either hinokiflavone, or DMSO, for 24 hr. HeLa, HEK293 and NB4 cells were examined for intron inclusion of HSP40, RIOK3, ACTR1b and DXO pre-mRNAs and for exon skipping of MCL1, NOP56, EIF4A2 and FAS pre-mRNAs. The positions of different cDNA products are pictured on the right of the gel images, and the molecular weight markers are shown on the left. Lane M, marker (hyperladder, 1 kb).
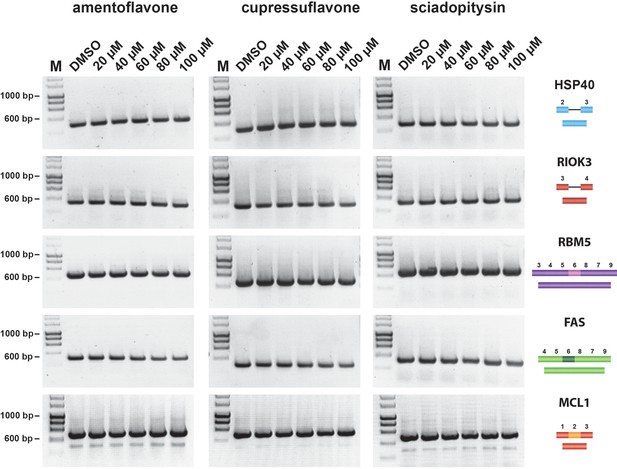
Amentoflavone, Cupressoflavone and Sciadopitysin do not alter pre-mRNA splicing in cellulo.
HEK293 cells were treated with either DMSO (control), or with 20 µM, 40 µM, 60 µM, 80 µM, or 100 µM of either amentoflavone, cupressoflavone, or sciadopitysin, for 24 hr. Using RT-PCR and primer pairs to detect changes in exon skipping of RBM5, FAS and MCL1 pre-mRNAs, as well as intron inclusion in HSP40 and RIOK3 pre-mRNAs, no alteration of pre-mRNA splicing for these transcripts were detected in the presence of these bioflavonoids. The positions of different cDNA products are pictured on the right of the gel images and the molecular weight markers are shown on the left. Lane M, marker (hyperladder, 1 kb).
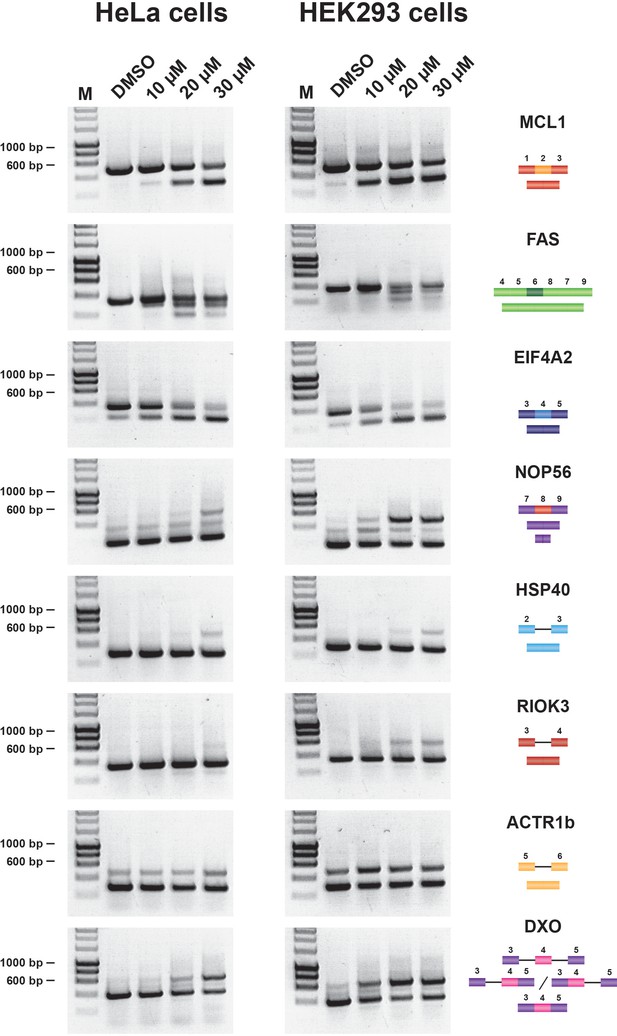
Isoginkgetin induces splicing changes in HeLa and HEK293 cells.
Semiquantitative RT-PCR analysis of cells treated with increasing concentrations of either hinokiflavone, or DMSO control, for 24 hr. HeLa and HEK293 were examined for intron inclusion of HSP40, RIOK3, ACTR1b and DXO pre-mRNAs and for exon skipping of MCL1, NOP56, EIF4A2 and FAS pre-mRNAs. The positions of different cDNA products are pictured on the right of the gel image and the molecular weight markers are shown on the left. Lane M, marker (hyperladder, 1 kb).
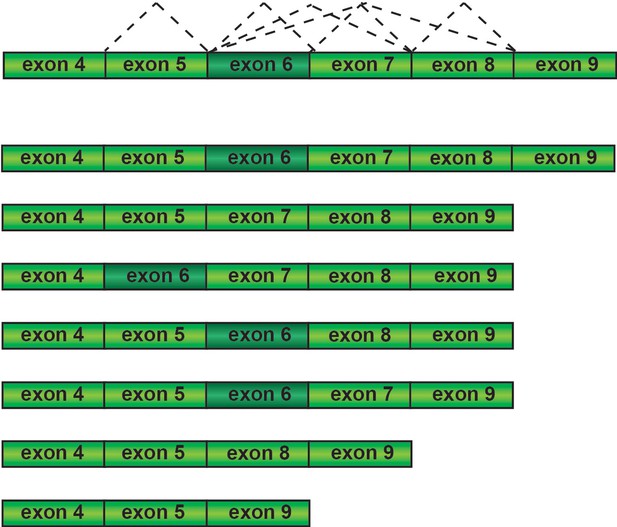
FAS pre-mRNA splicing in the presence of hinokiflavone.
Schematic representation of FAS isoforms formed in hinokiflavone treated cells.
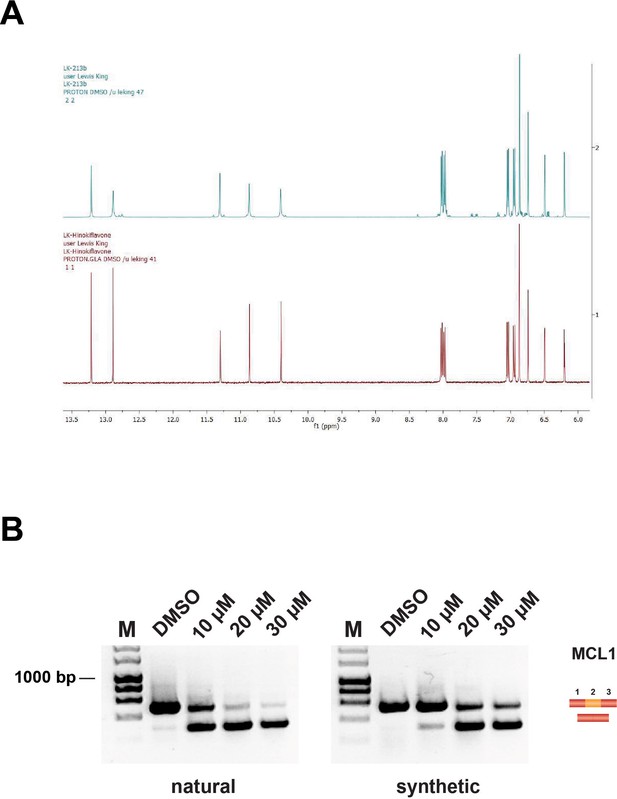
Comparison of natural and synthetic hinokiflavone.
(A) Proton nuclear magnetic resonance spectrum of natural (red) and synthetic (blue) hinokiflavone. (B) Effect of natural and synthetic hinokiflavone on the alternative splicing of the MCL1 pre-mRNA in HEK293 cells.
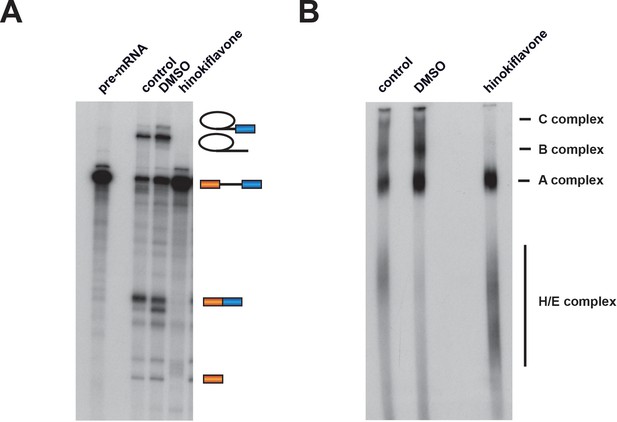
Hinokiflavone blocks spliceosome assembly prior to B complex formation.
Formation of splicing complexes on the Ad1 pre-mRNA was analysed on a native agarose gel after incubation with either DMSO (control), or 500 µM hinokiflavone. The positions of the splicing complexes C, B, A and H/E are indicated on the right.
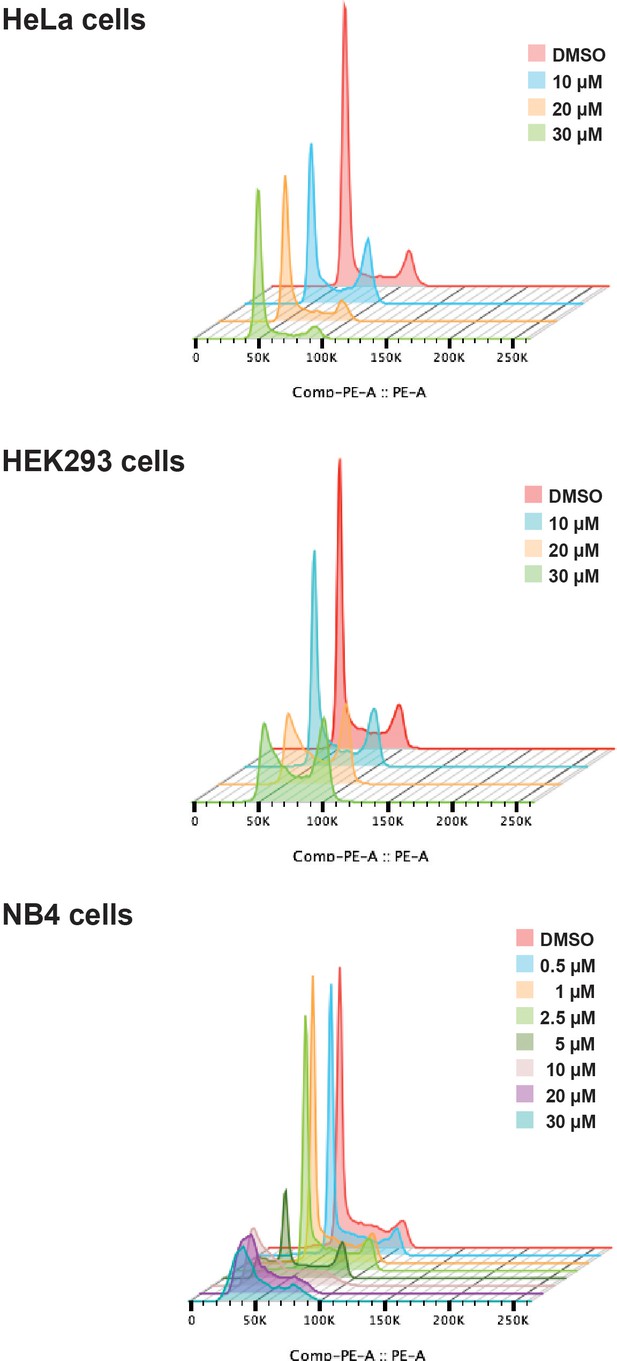
Hinokiflavone shows cell cycle specific effects.
Cell cycle analysis was performed on HeLa, HEK293 and NB4 cells treated with either different concentrations of hinokiflavone, or DMSO (control), for 24 hr. Cellular DNA content was measured by propodium iodide staining followed by flow cytometry analysis.
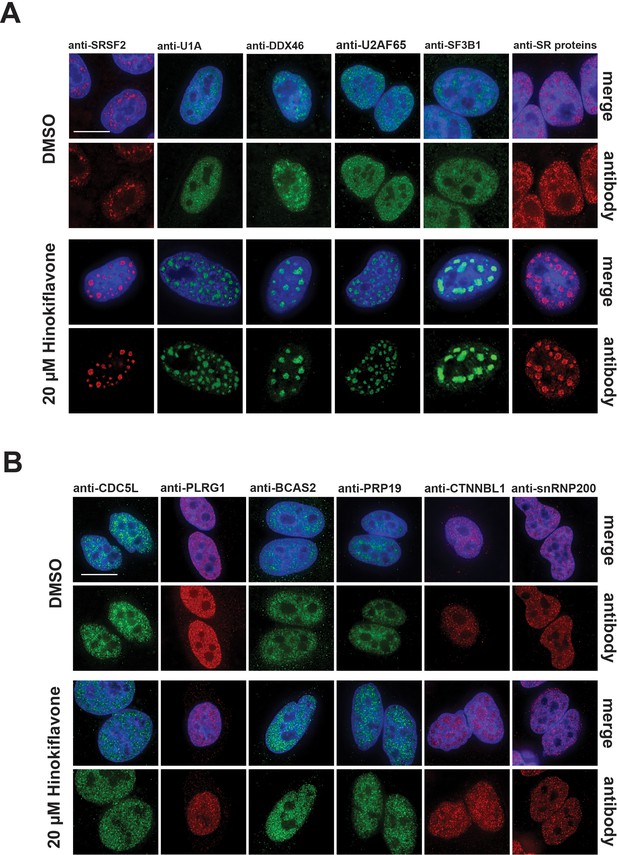
Changes in splicing speckles after treatment with hinokiflavone.
HeLa cells incubated for 24 hr with either DMSO (control), or 20 µM hinokiflavone, were fixed and stained with antibodies for the following splicing factors: SRSF2, U1A, U2AF65, DDX46, SF3B1, SR proteins, CDC5L, PLRG1, BCAS2, PRP19, snRNP200 and CTNNBL1. (A) Splicing factors involved in the early steps of spliceosome assembly are located in enlarged splicing speckles. (B) Splicing factors that assemble after A complex formation are not accumulated in enlarged speckles and show a more diffuse nucleoplasmic distribution. Scale bars, 15 µm.
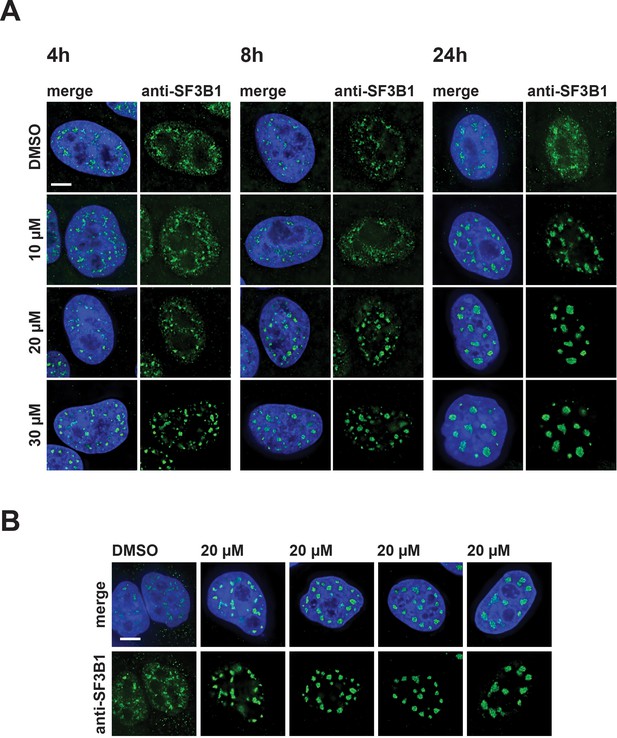
Mega speckles vary in size and number.
(A) HeLa cells incubated for 4 hr, 8 hr, or 24 hr, with either DMSO (control), or 10 µM, 20 µM, or 30 µM hinokiflavone, were fixed and stained with an antibody against SF3B1. A dose and time dependent increase in the size of mega speckles can be observed. (B) The numbers of mega speckles at 20 µM hinokiflavone after 24 hr of treatment varies from over 30, to less than 10, per cell. Their size also varies from ~0.5 µm to over 4 µm. Scale bars, 6.5 µm.
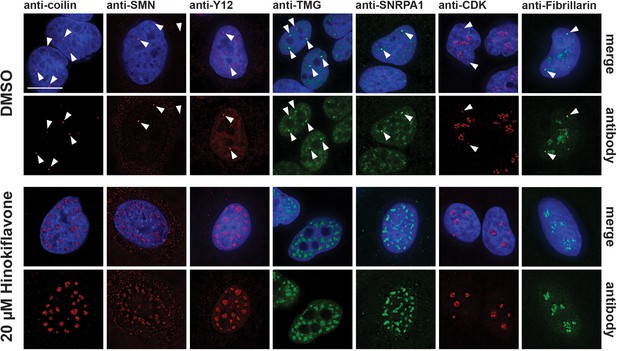
Hinokiflavone treatment leads to relocation of CB components to mega speckles.
HeLa cells were treated for 24 hr with either DMSO (control), or 20 µM hinokiflavone and the fixed cells were stained with anti-coilin, anti-SMN, anti-Y12, anti-TMG, anti-SNRPA1, anti-CDK and anti-Fibrillarin antibodies, respectively. Coilin, SMN, Y12 and TMG show relocation to the enlarged speckles containing splicing factors in hinokiflavone treated cells. Arrowheads denote intact CBs. CDK and Fibrillarin are only detected in nucleoli after treatment with hinokiflavone. Scale bars, 15 µm.
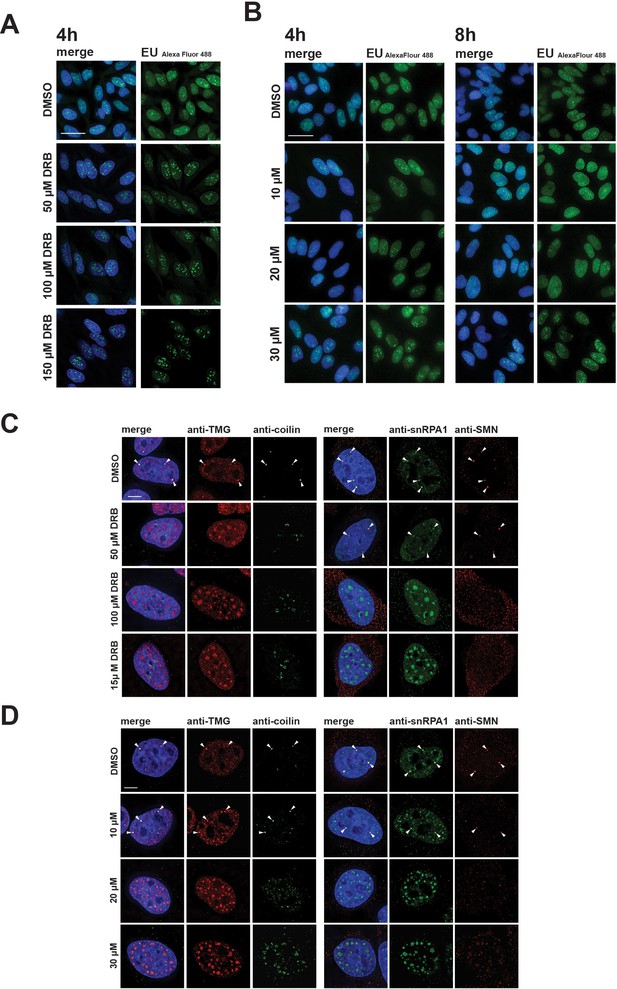
Differential effects of hinokiflavone and DRB on inhibition of RNA polymerase II transcription and disruption of Cajal bodies.
HeLa cells were incubated for either 4 hr with DRB (A), or 4 hr and 8 hr with hinokiflavone (B), before cells were incubated with EU to label newly synthesized RNA. Cells were fixed and labelled RNA was detected by fluorescence microscopy. Scale bar, 40 µm. (C) HeLa cells were incubated with either DMSO (control), or with 50 µM, 100 µM, or 150 µM DRB, for 4 hr, then stained with anti-coilin, anti-TMG, anti-SMN and anti-SNRPA1 antibodies, respectively. Arrowheads denote intact CBs. (D) HeLa cells were incubated with either DMSO (control), or with 10 µM, 20 µM, or 30 µM hinokiflavone, for 8 hr, then fixed and stained with anti-coilin, anti-TMG, anti-SMN and anti-SNRPA1 antibodies, respectively. Arrowheads denote intact CBs. Scale bar, 6.5 µM.
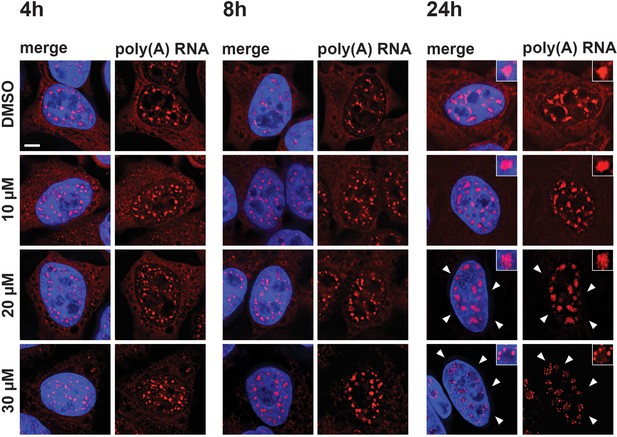
Hinokiflavone affects localization of polyadenylated RNA.
HeLa cells were treated for 4 hr, 8 hr, or 24 hr, with either DMSO (control), or 10 µM, 20 µM, or 30 µM hinokiflavone and the fixed cells were hybridized with Cy3 labelled Oligo dT probes. After 24 hr the poly(A) RNA is lost from the cytoplasm (arrowheads) in the presence of 20 and 30 µM hinokiflavone. Treatment with 30 µM hinokiflavone leads to a relocation of poly(A) RNA from the enlarged splicing speckles to circles of dots. Scale bars, 6.5 µm.
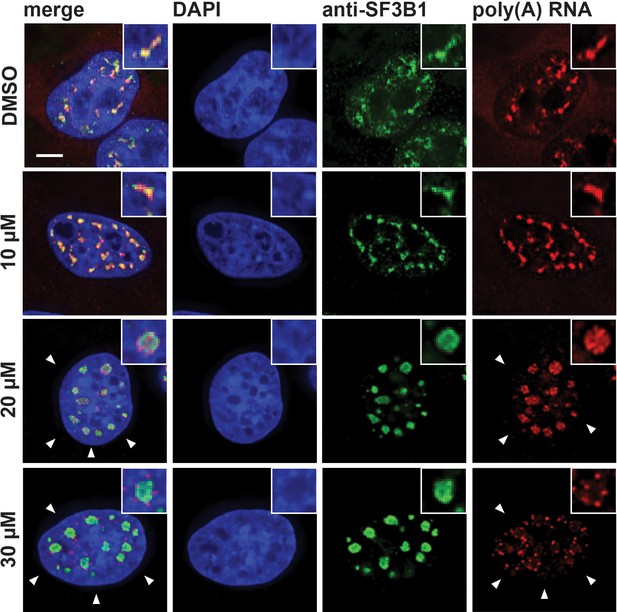
Hinokiflavone affects localization of polyadenylated RNA.
HeLa cells were incubated for 24 hr with either DMSO, or 10 µM, 20 µM, or 30 µM hinokiflavone and the fixed cells were hybridized with Cy3 labelled Oligo dT probes and co-stained with anti-SF3B1. Treatment with 30 µM hinokiflavone leads to a relocation of polyadenylated RNA from the enlarged splicing speckles to dots at their periphery. Scale bars, 6.5 µm.
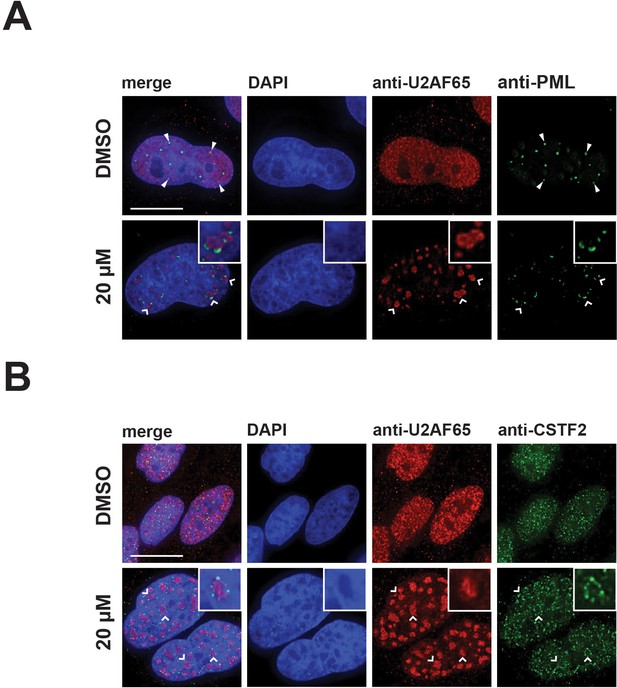
Hinokiflavone treatment leads to the relocation of nuclear proteins to the periphery of enlarged splicing speckles.
HeLa cells were treated for 24 hr with either DMSO (control), or 20 µM hinokiflavone and the fixed cells were stained with either (A) anti-U2AF65 and anti-PML, or (B) anti-CSTF2 antibodies. Co-staining with anti-U2AF65 antibodies showed that both CSTF2 and PML relocate to the periphery of enlarged splicing speckles (highlighted by carets and enlarged images). Arrowheads denote PML bodies. Scale bars, 15 µm.
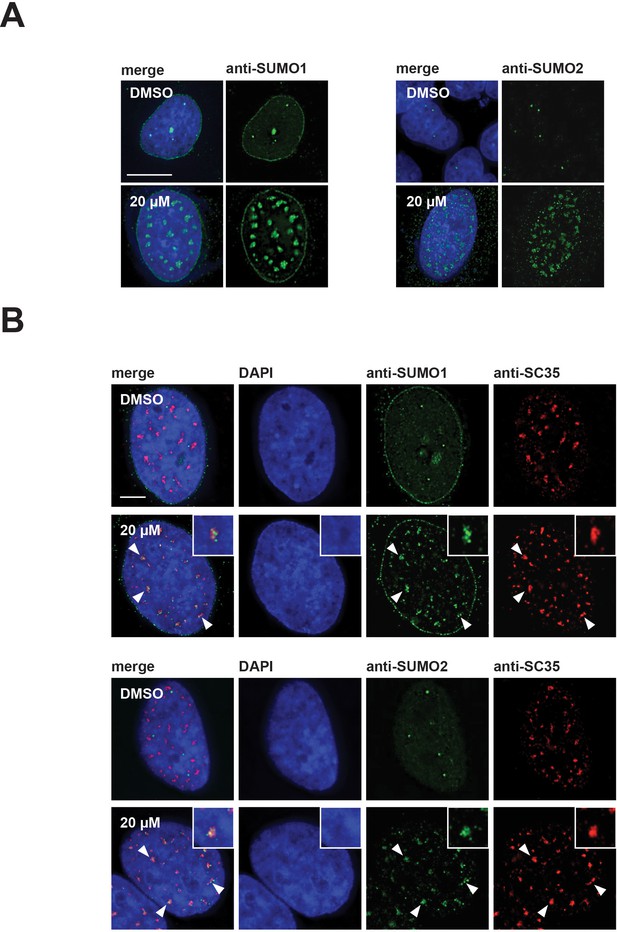
SUMO1 and SUMO2/3 relocalize to enlarged splicing speckles in the presence of hinokiflavone.
(A) HeLa cells were treated for 24 hr with either DMSO (control), or 20 µM hinokiflavone and the fixed cells were stained with either anti-SUMO1, or anti-SUMO2/3 antibodies. Both SUMO1 and SUMO2/3 accumulated in the enlarged splicing speckles formed after treatment with hinokiflavone. (B) Treatment of HeLa cells with 20 µM hinokiflavone for 2 hr. Co-staining with either anti-SUMO1, or anti-SUMO2/3 and anti-SRSF2 (SC35) antibodies showed that both SUMO1 and SUMO2/3 accumulate in enlarged splicing speckles (highlighted by arrowheads and enlarged images). Scale bars, 15 µm.
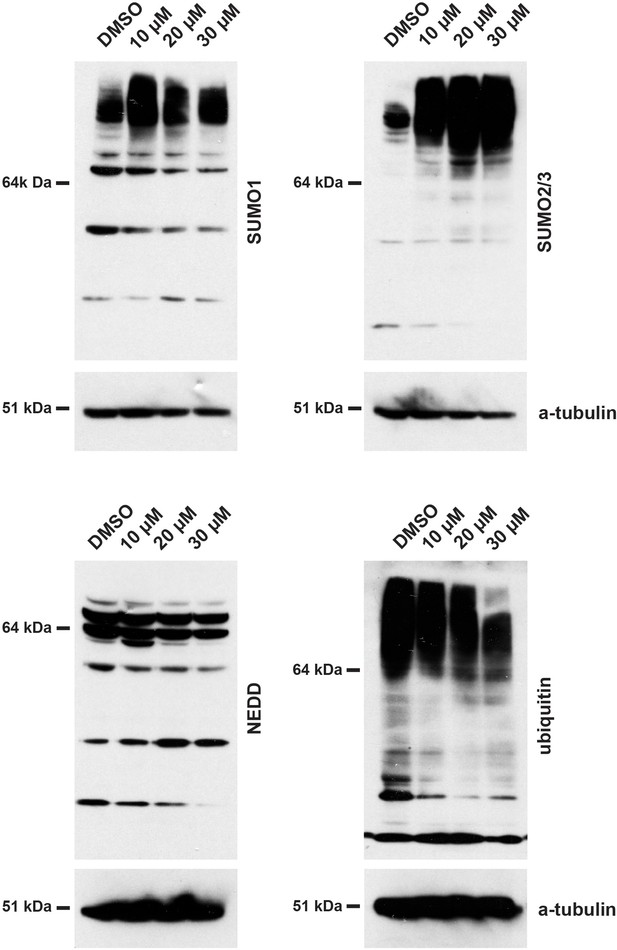
Treatment of HEK293 cells with hinokiflavone leads to an increase in SUMOylated proteins.
HEK293 cells were treated with either DMSO (control), or with 10 µM, 20 µM, or 30 µM hinokiflavone for 24 hr before cells were lysed in 1x LDS buffer. Samples were separated on SDS-PAGE and transferred to membranes. After probing with antibodies specific for SUMO1, SUMO2/3, Ubiquitin, and NEDD8, labelled proteins were visualized using chemiluminescence, showing a specific accumulation of poly-SUMOylated proteins after hinokiflavone treatment. The membranes were also probed to detect alpha-tubulin as a control (bottom panels).
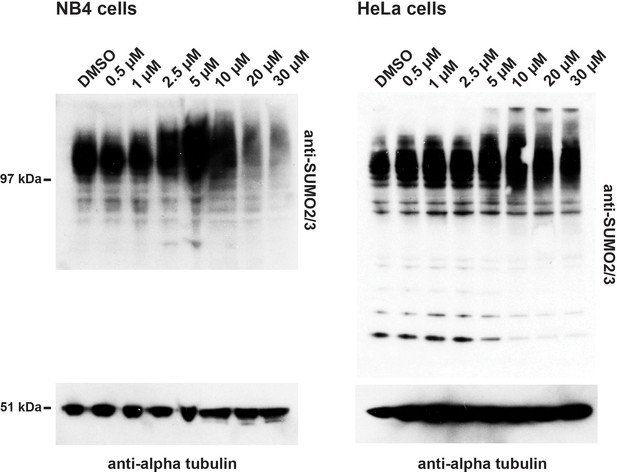
Hinokiflavone treatment leads to an increase in SUMO2/3 modified proteins in HeLa and NB4 cells.
HeLa and NB4 cells were each treated with either DMSO (control), or with 0.5 µM, 1 µM, 2.5 µM, 5 µM, 10 µM, 20 µM, or 30 µM hinokiflavone for 24 hr. SUMO2/3 and alpha tubulin were detected by western blotting.
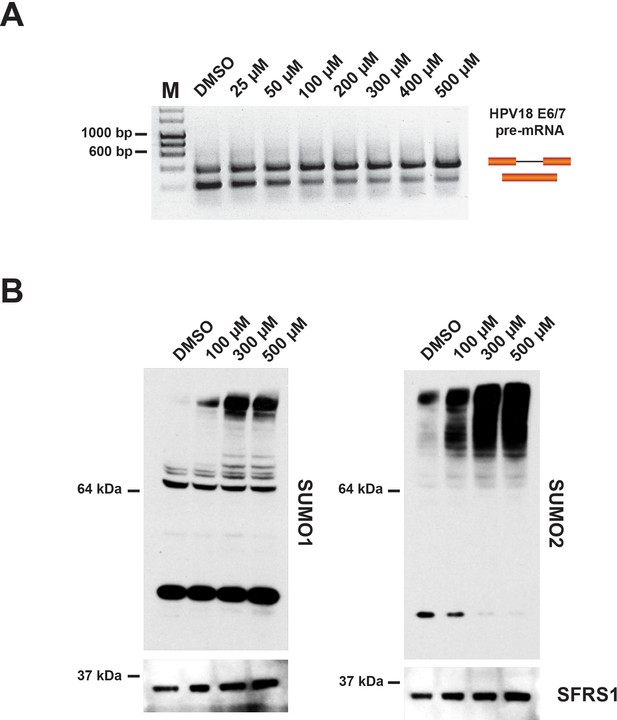
Incubation of in vitro nuclear splicing extracts with hinokiflavone leads to an increase in SUMOylated proteins.
HeLa nuclear extract reactions were incubated in vitro under splicing conditions with either DMSO (control), or with increasing concentrations of hinokiflavone from 25 µM-500 µM. (A) Evaluation of the lowest concentration at which hinokiflavone inhibits pre-mRNA splicing in vitro. (B) Proteins were extracted and size-separated by SDS-PAGE, transferred to membranes and probed using either anti-SUMO1, anti-SUMO2/3, or anti-SRSF1 antibodies and visualized using chemiluminescence. A specific accumulation of hyper-SUMOylated proteins after hinokiflavone treatment is shown. Membranes were also probed to detect SFRS1 as a loading control (bottom panels).
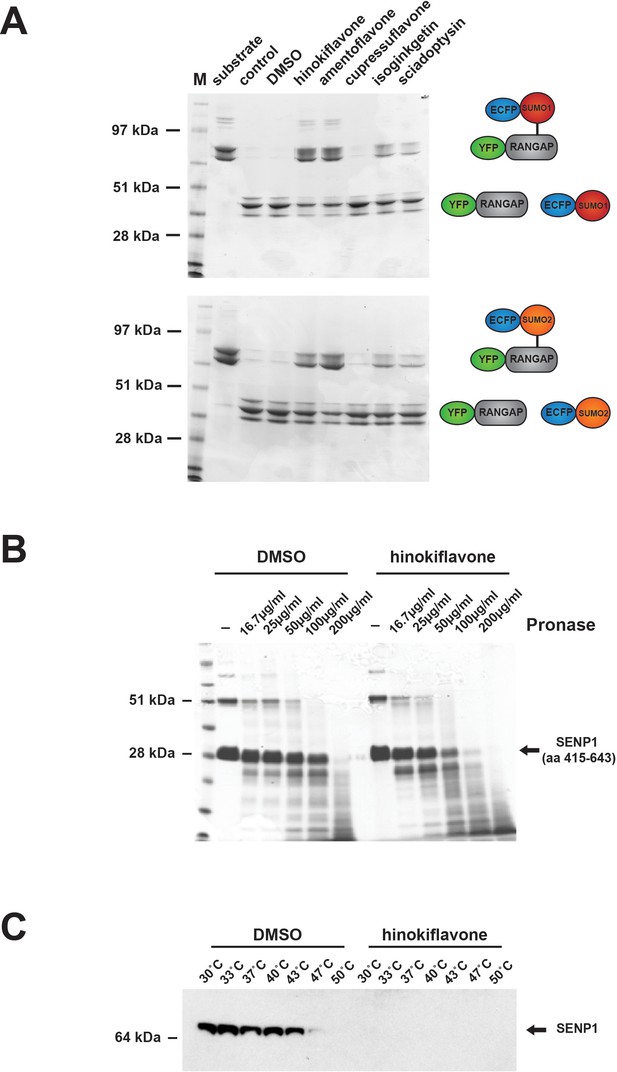
Biflavones inhibit SENP1 in vitro.
(A) The effect of 500 µM hinokiflavone, amentoflavone, cupressuflavone, isoginkgetin and sciadopitysin on the isopeptidase activity of a highly purified fragment of catalytically active SENP1 (comprising aa 415–643), was determined by an in vitro gel-based activity assay. (B) DARTS Assay; incubation of the catalytically active SENP1 fragment with either DMSO (control), or 500 µM hinokiflavone, before the samples were digested with different concentrations (from ~16–200 µg/ml) of the proteinase pronase. SENP1 showed an increased sensitivity to protease digestion in the presence of hinokiflavone. (C) CESTA Assay; HeLa nuclear extract was treated with either DMSO, or 500 µM hinokiflavone for 20 min at RT, followed by heat treatment and ultracentrifugation. Western blot analysis of the soluble proteins demonstrated a dramatic change in the thermal stability of SENP1 in the presence of hinokiflavone.
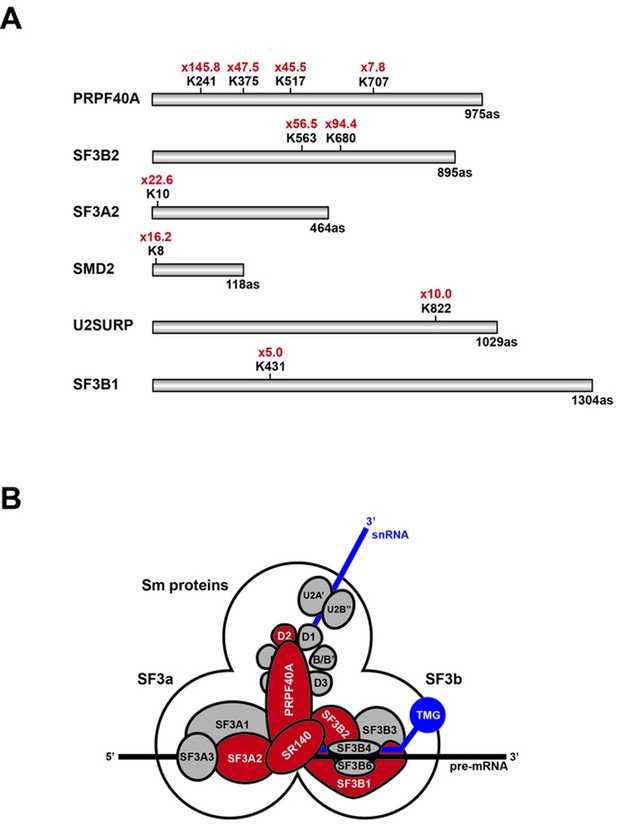
Schematic representation of splicing factors identified as SUMO2 target proteins.
(A) Lysine residues in PRPF40A, SF3B2, SF3A2, SMD2, SR140 (U2SURP) and SF3B1 that show increased SUMO2 modifications in HEK293 cells treated for 8 hr with 20 µM hinokiflavone are shown. (B) Schematic representation of the SUMO2-modified U2 snRNP components, which are coloured in red.
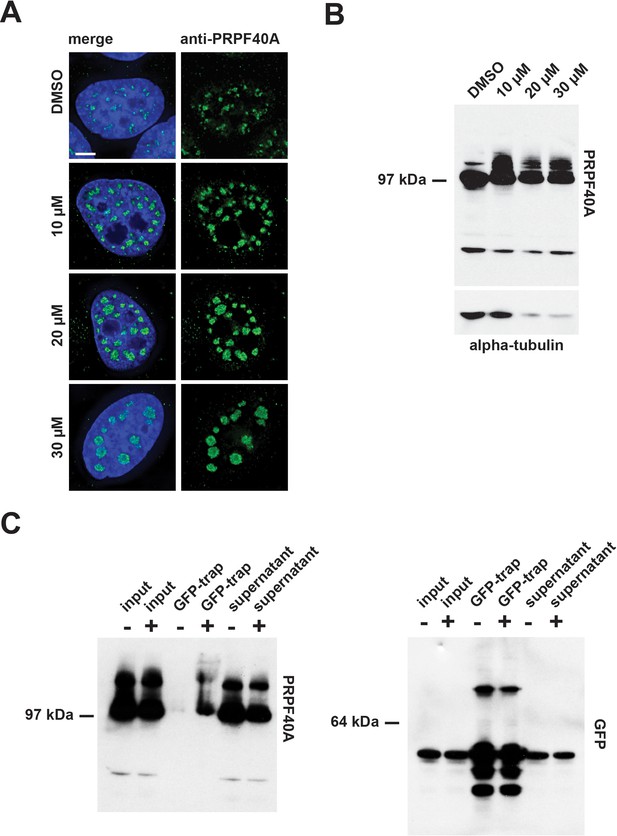
Confirmation of PRPF40A as a SUMO target protein.
(A) Immunofluorescence analysis shows that hinokiflavone treatment leads to the relocation of PRPF40A to mega speckles in HeLa cells. Scale bar represents 6.5 µm. (B) HEK293 cells were treated with either DMSO (control), or with 10 µM, 20 µM, or 30 µM hinokiflavone, for 24 hr, then total cell lysate proteins were size-separated by SDS-PAGE, transferred to membranes, probed using the anti-PRPF40A antibody and visualized using chemiluminescence. In the presence of hinokiflavone additional higher molecular weight bands are detected. (C) YFP-SUMO2 expressing HeLa cells were treated either with DMSO (-), or 20 µM hinokiflavone (+), for 8 hr. Cells were lysed and the YFP-SUMOylated proteins immunoprecipitated with GFP-trap beads. The input, IPs, pellets and unbound fractions of both the control and hinokiflavone treated cells were size separated by SDS-PAGE, transferred to membranes and probed using anti-PRPF40A and anti-GFP antibodies and visualized using chemiluminescence.
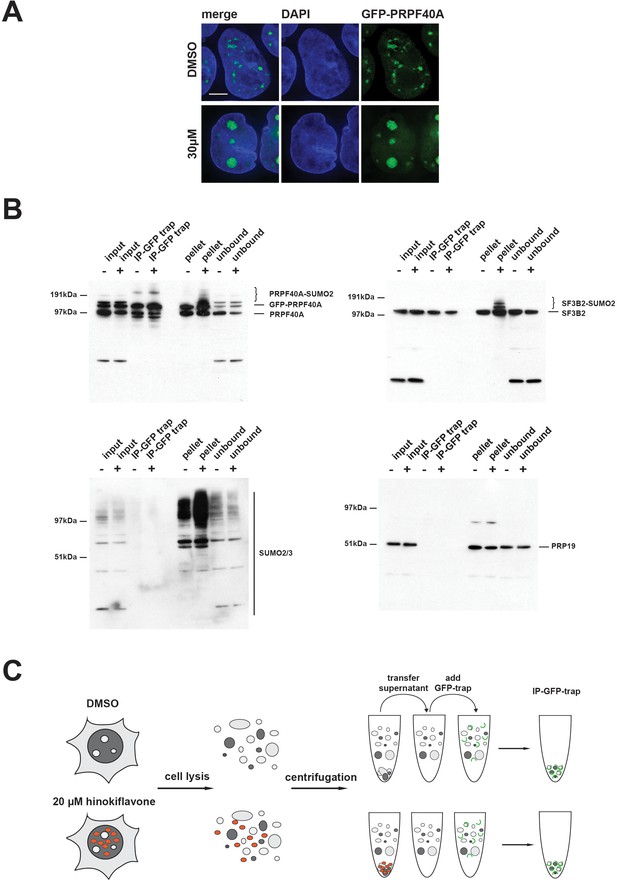
SUMOylated PRPF40A and SF3B2 accumulate in the insoluble fraction of HEK293 cell lysates.
(A) GFP-PRPF40A stably expressed in HEK293 cells relocates to mega speckles in the presence of 30 µM hinokiflavone, as shown by fluorescence microscopy analysis. Scale bar represents 6.5 µM. (B) HEK293 cells stably expressing GFP-PRPF40A were treated with either DMSO (-) as a negative control, or with 20 µM hinokiflavone (+), for 8 hr. Cells were lysed and analyzed by co-immunoprecipitation (Co-IP). The input, IPs, pellets and unbound fractions of both the control and hinokiflavone treated cells were size separated by SDS-PAGE, transferred to membranes and probed using anti-PRPF40A, anti-SF3B2, anti-SUMO2/3 and anti-PRP19 antibodies and visualized using chemiluminescence. (C) Schematic representation of the Co-IP procedure.
Tables
SUMO-modified lysine residues in U2 snRNP proteins upregulated by hinokiflavone.
https://doi.org/10.7554/eLife.27402.023| Gene name | Modified sequence | Position | Upregulation |
|---|---|---|---|
| PRPF40A | _SNLHAM(ox)IK(gl)AEESSK_ | 241 | 145.8 |
| _DVLFFLSK(gl)K_ | 517 | 47.5 | |
| _TVADFTPK(gl)K_ | 375 | 45.5 | |
| _DFVAIISSTK(gl)RSTTLD_ | 707 | 7.8 | |
| SF3B2 | _TGK(gl)PLYGDVFGTNAAE_ | 680 | 94.4 |
| _M(ox)GK(gl)IDIDYQK_ | 563 | 56.5 | |
| SF3A2 | _(ac)M(ox)DFQHRPGGK(gl)TGSGGVASSSE_ | 10 | 22.6 |
| SNRPD2 | _PK(gl)SEM(ox)TPEELQK_ | 8 | 16.2 |
| U2SURP | _HHLYSNPIK(gl)EE_ | 822 | 10.0 |
| SF3B1 | _GYK(gl)VLPPPAGYVPIRTPARK_ | 413 | 5.0 |
Primary antibodies used for IF staining and/or western-blotting.
https://doi.org/10.7554/eLife.27402.026| Primary antibody | Company |
|---|---|
| anti-BCAS2 | Abcam, Cambridge, UK (RRID:AB_1861326) |
| anti-CDC5L | BD Transduction Laboratories (RRID:AB_399724) |
| anti-coilin | Proteintech, Chicago, IL, US (RRID:AB_2276345) |
| anti-CSTF2 | Santa Cruz, US (RRID:AB_668179) |
| anti-CTNNBL1 | Abcam, Cambridge, UK (RRID:AB_1523420) |
| anti-DDX46 | Proteintech, Chicago, IL, US (RRID:AB_2090927) |
| anti-Fibrillarin | Cytoskeleton Inc, Denver, CO, US (RRID:AB_10709399) |
| anti-PRPF40A | Novus, Biologicals, Chambridge, UK (RRID:AB_11012473) |
| anti-PLRG1 | Abcam, Cambridge, UK (RRID:AB_2170868) |
| anti-PRP19 | Abcam, Cambridge, UK (RRID:AB_2170868) |
| anti-NEDD8 | Cell Signaling Technology, Danvers, MA, US (RRID:AB_659972) |
| anti-SF3B1 | Abcam, Cambridge, UK (RRID:AB_2186512) |
| anti-SF3B2 | Novus Biologicals, Abington, UK (RRID:AB_1110397) |
| anti-SFRS1 | Abcam, Cambridge, UK (RRID:AB_298608) |
| anti-SMN | ImmunoGlobe, Himmelstadt, Germany (RRID:AB_2687973) |
| anti-SNRPA1 | Abcam, Cambridge, UK (RRID:AB_11139816) |
| anti-snRNP200 | Abcam, Cambridge, UK (RRID:AB_10901078) |
| anti-SR proteins | Merck Group, Darmstadt, Germany (RRID:AB_10807429) |
| anti-SUMO1 | Cell Signaling Technology, Danvers, US (RRID:AB_10698887) |
| anti-SUMO2/3 | Cell Signaling Technology, Danvers, US (RRID:AB_2198425) |
| anti-TMG | Merck Group, Darmstadt, Germany (RRID:AB_2687977) |
| anti-U1A | Iain Mattaj (RRID:AB_2713922) |
| anti-U2AF65 | SIGMA, ST Louis, Missouri, US (RRID:AB_262122) |
| anti-Ubiquitin | Cell Signaling Technology, Danvers, US (RRID:AB_2180538) |
| anti-Y12 | Joan Steitz (RRID:AB_2692320) |
Primer pairs used for RT-PCR to identify pre-mRNA splicing changes.
https://doi.org/10.7554/eLife.27402.027| Primer | Sequence |
|---|---|
| ACTR1b for | CCGCTCAACCCGAGTAAGAA |
| ACTR1b rev | CAGCCGAGGTATGGAAGTCA |
| DXO for | TGGGGAGGTTAACACCAACG |
| DXO rev | GCTCTGGGAAAGCTAAGGA |
| EIF4A2 for | GTCTCTCCTTCGTGGCATCT |
| EIF4A2 rev | TCTCCCGGGTGTACCAACA |
| HSP40 for | GAACCAAAATCACTTTCCCCAAGGAAGG |
| HSP40 rev | AATGAGGTCCCCACGTTTCTCGGGTGT |
| MCL1 for | GAGGAGGAGGAGGACGAGTT |
| MCL1 rev | ACCAGCTCCTACTCCAGCAA |
| NOP56 for | GCATCCACAGTGCAGATCCT |
| NOP56 rev | GCAATCGATTCGTGAGGCAA |
| FAS for | CCCGGCCCAGAAATACCAAG |
| FAS rev | GACTCCAGCAATAGTGGTGATA |
| RIOK3 for | GCTGAAGGACCATTTATTACTGGAG |
| RIOK3 rev | TTCTTGCTGTGTTCTTTCTCCCACAC |
Additional files
-
Supplementary file 1
List of lysine residues identified to show an increase in SUMO2-modification in HEK293 cells 8 hr after treatment with 20 µM hinokiflavone.
Corresponding gene and protein names are indicated for each lysine.
- https://doi.org/10.7554/eLife.27402.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.27402.029





