Multiplexed genetic engineering of human hematopoietic stem and progenitor cells using CRISPR/Cas9 and AAV6
Figures
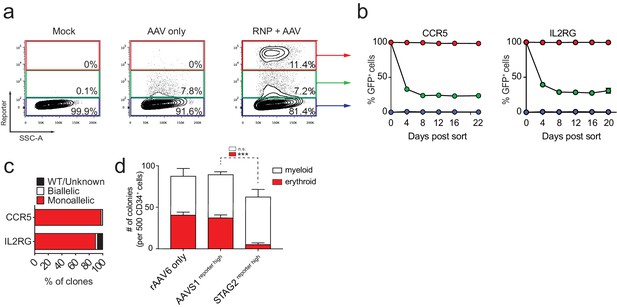
FACS-based identification and enrichment of monogenic genome-edited CD34+ human hematopoietic stem and progenitor cells (HSPCs).
(a) HSPCs were electroporated with CCR5-RNP and transduced with CCR5-tNGFR rAAV6 HR donor. Representative FACS plots from day four post-electroporation highlight the CCR5 tNGFRhigh population (red gate) generated by the addition of Cas9 RNP compared to cells with low reporter expression (green gate) and reporternegative cells (black gate). Numbers reflect percentage of cells within gates. (b) Day four post-electroporation, CCR5 (tNGFR or GFP) and IL2RG (GFP)-targeted HSPCs from reporterhigh (red), reporterlow (green), and reporterneg (blue) fractions were sorted and cultured for 20-22 days while monitoring the percentage of cells that remained GFP+. Error bars represent S.E.M. N = 6 for CCR5, N = 3 for IL2RG, all from different CD34+ donors. (c) HSPCs were targeted at CCR5 (with GFP or tNGFR donor) or at IL2RG (GFP donor; only female cells for IL2RG). At day four post-electroporation, reporterhigh cells were single-cell sorted into methylcellulose for colony formation. PCR was performed on colony-derived gDNA to detect targeted integrations. 338 CCR5 and 177 IL2RG myeloid and erythroid methylcellulose colonies were screened from at least two different CD34+ HSPC donors. (d) HSPCs were targeted at the STAG2 gene or the AAVS1 locus with a GFP reporter cassette. Cells that only received the STAG2-GFP AAV6 donor and not Cas9 RNP were included as an additional control. At day four post-electroporation and transduction, reporterhigh cells from the STAG2 and AAVS1 targeting experiments and bulk cells from the STAG2 AAV6 only population were plated in methylcellulose for colony formation. After 14 days, colonies were scored as either erythroid or myeloid based on morphology. Error bars represent S.E.M, N = 3, ***p<0.001, n.s. = p≥0.05, unpaired t-test.
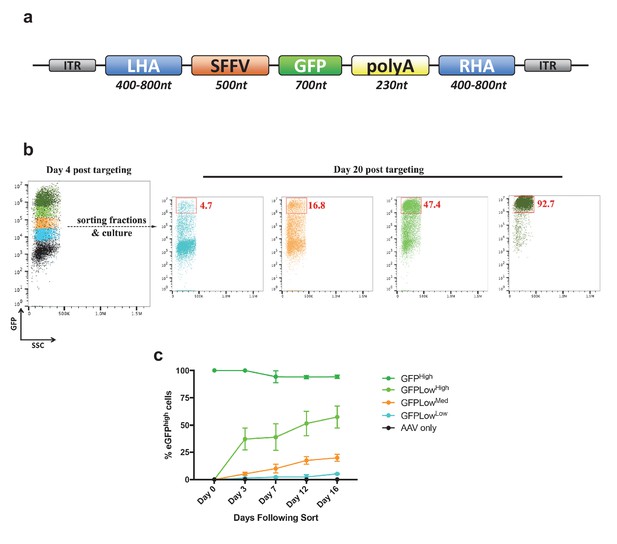
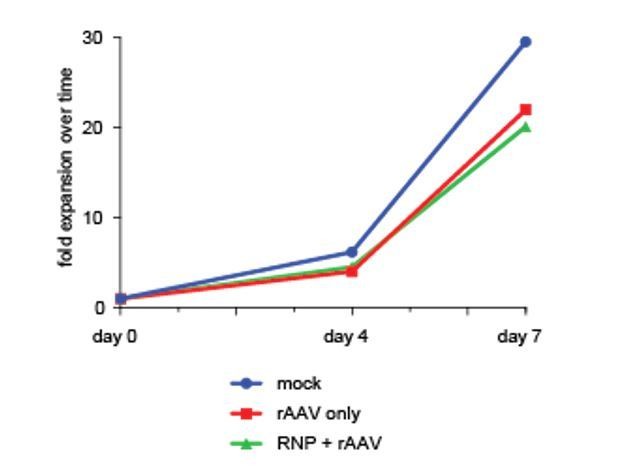
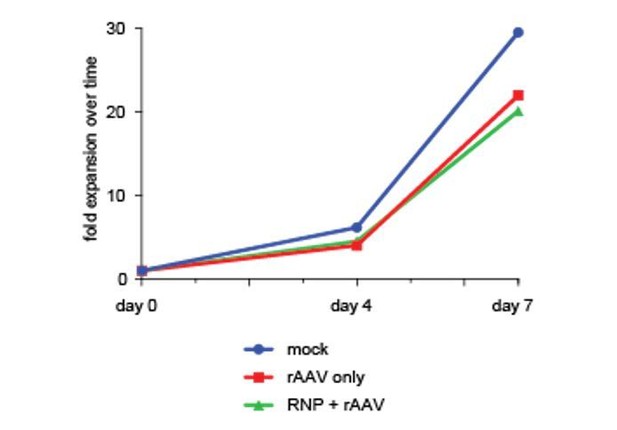
Analysis of cell fractions with different fluorescence intensity.
(a) Schematic showing the general layout of the AAV6 donors employed. ITR: inverted terminal repeat; SFFV promoter: spleen focus forming virus promoter; GFP: green fluorescent protein; polyA: bovine growth hormone polyadenylation signal; RHA: right homology arm. Approximate sizes are shown below each component. (b) Cells were targeted at the HBB locus by electroporation of Cas9 RNP followed by transduction of a homologous rAAV6 donor carrying a GFP expression cassette. At 4 days post electroporation and transduction, cells with different GFP intensities (GFPhigh, GFPLowHigh, GFPLowMed, GFPLowLow) were FACS-sorted and cultured for an additional 16 days. At day 20 post targeting, cells were analyzed for GFP expression by flow cytometry and the red gates show the GFPhigh population at this time point. (c) The cells from b) were analyzed at different time points after sorting, and data points show the percentage of cells within the GFPhigh gate for the different populations as well as a population receiving only the rAAV6 donor and not Cas9 RNP.
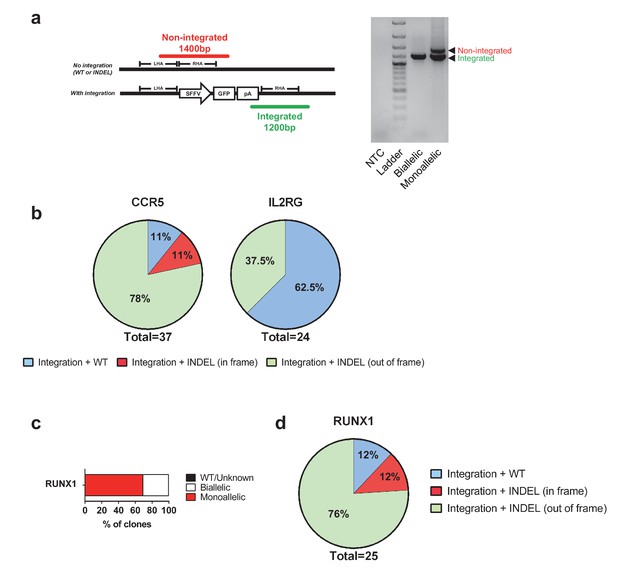
Genotypes of clones with mono-genic targeting.
(a) Left, schematic representation of the three-primer PCR used to genotype CCR5 alleles for integrated (green PCR product) and non-integrated (red PCR product) alleles. One forward primer is located in the left homology arm (LHA), one forward primer is located in the poly A, and a common reverse primer is located outside the region of the right homology arm (RHA). Right, gel image of representative genotyped clones from Figure 1c (CCR5) showing colonies with biallelic and monoallelic integrations. (b) A subset of the CCR5 and IL2RG clones (only female cells for IL2RG) from Figure 1c with monoallelic integration had the genotype on the non-integrated allele analyzed by Sanger sequencing of purified PCR products. Note that in-frame INDELs can be gene-disrupting depending on the location and size of the INDEL. (c) As in Figure 1c, HSPCs were targeted at RUNX1 and at day four post-electroporation, reporterhigh cells were single cell-sorted into methylcellulose-containing 96-well plates to establish colonies. After 14 days, PCR was performed on colony-derived gDNA to detect targeted integrations. A total of 36 myeloid and erythroid methylcellulose colonies were screened. (d) The monoallelically targeted clones from c) had the genotype assessed on the non-integrated allele by Sanger sequencing of purified PCR products. See Supplementary file 1b for complete list of genotypes.
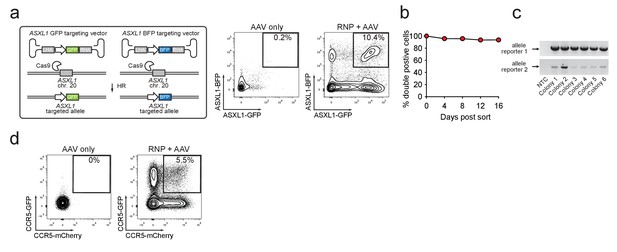
Identification and enrichment of biallelic genome-edited CD34+ human hematopoietic stem and progenitor cells (HSPCs).
(a) Left, Schematic showing biallelic targeting strategy for ASXL1 using GFP and BFP-encoding rAAV6 donors for integration into each allele of ASXL1. The SFFV promoter drives reporter expression. Middle, FACS plot from an ‘AAV only’ sample day four post electroporation, showing low episomal reporter expression (BFP and GFP) in cells without the CRISPR system. Right, FACS plot of CD34+ HSPCs treated with both Cas9 RNP and the two rAAV6 donors highlighting the generation of BFPhigh/GFPhigh double positive cells that have undergone ASXL1 dual-allelic targeting. (b) HSPCs were targeted at both alleles of HBB (Cas9 RNP with GFP and tdTomato rAAV6 donors) and at day four post electroporation, dual positive cells were sorted and cultured for 16 days while analyzing reporter expression. Error bars representing S.E.M. are present, but too small to be visible (N = 3 different HSPC donors). (c) Gel images showing PCR genotyping of six methylcellulose-derived clones from (e) confirming integration into each of the HBB alleles. (d) Human primary T cells were CD3/CD28 stimulated for three days and then electroporated with CCR5-targeting Cas9 RNP and transduced with two CCR5-specific rAAV6 donors encoding GFP and mCherry, respectively. FACS plots show GFPhigh/mCherryhigh biallelic targeting frequencies at day four post-electroporation.
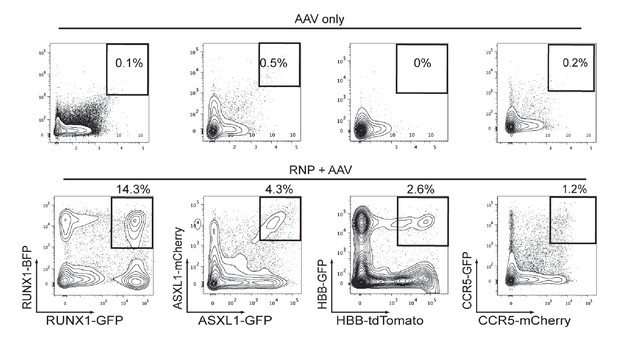
Cas9 and rAAV6-mediated biallelic homologous recombination (HR) in human CD34+HSPCs.
Top, Representative FACS plots from HSPCs transduced with two rAAV6 (two fluorescent reporters for each gene), that have homology for the genes listed on the bottom panel, show low episomal expression and very few dual reporterhigh-expressing HSPCs. Bottom, HSPCs were electroporated with gene-specific Cas9 RNPs and then transduced with rAAV6 targeting each allele of a gene with two different indicated fluorescent reporters. FACS plots at Day 4-post electroporation highlight the dual reporterhigh cells that have undergone HR at both alleles of the intended gene.
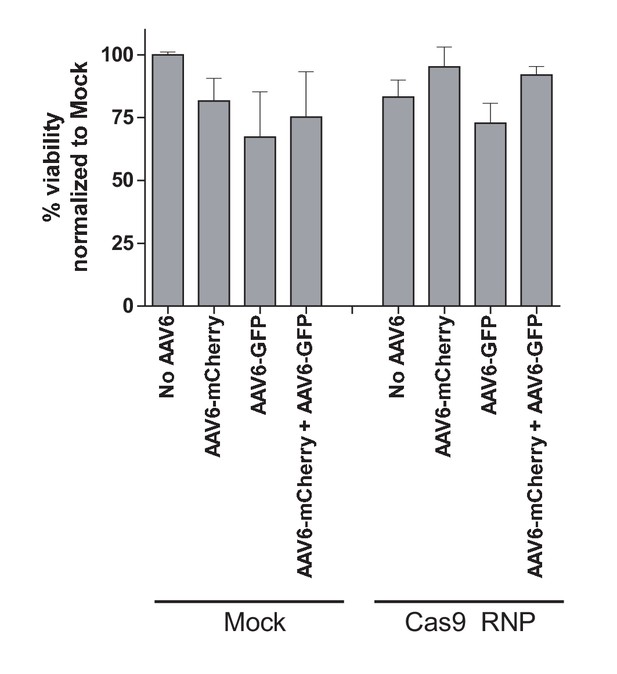
Toxicity assessment of biallelic integration at the CCR5 locus in primary human T cells.
Human primary T cells were isolated from buffy coats and stimulated for three days using anti-CD3 and anti-CD28 antibodies. Cells were then electroporated with CCR5-targeting Cas9 RNP and transduced with two CCR5-specific rAAV6 donors encoding GFP and mCherry, respectively, either alone or in combination. Cell viabilities were measured at Day two post-electroporation by Trypan Blue exclusion assay (N = 2 different buffy coat-derived T cells).
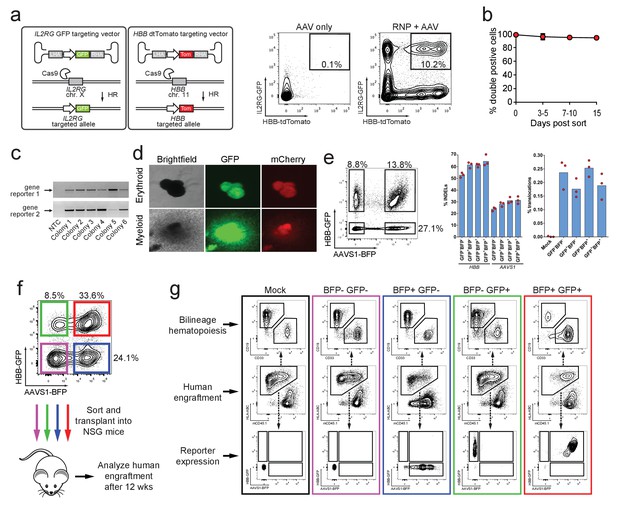
Identification, enrichment, and long-term engraftment in NSG mice of di-genic genome-edited CD34+ human hematopoietic stem and progenitor cells (HSPCs).
(a) Left, Schematic depicting HBB and IL2RG di-genic targeting. Middle, FACS plot of an ‘AAV only’ sample at day four post electroporation, showing low episomal reporter expression (HBB-tdTomato and IL2RG-GFP) in cells without the CRISPR system. Right, FACS plot at day four post-electroporation of HSPCs electroporated with Cas9 RNP targeting both HBB and IL2RG followed by transduction with HBB-tdTomato and IL2RG-GFP rAAV6 donors showing the generation of tdTomatohigh/GFPhigh cells with di-genic targeting at HBB and IL2RG. (b) Double-positive HSPCs targeted at HBB (GFP) and CCR5 (mCherry) were sorted at day four post-electroporation and cultured for 15 days while analyzing reporter expression. Error bars represent S.E.M. (N = 3 different HSPC donors). (c) Representative gel images showing PCR genotyping of six (out of 57 total) HBB-GFPhigh (gene reporter 1)/CCR5-mCherryhigh (gene reporter 2) methylcellulose-derived clones confirming integration at each locus (d) Representative fluorescence microscopy images of methylcellulose-derived clones with di-genic targeting at HBB and CCR5 show myeloid and erythroid progenitors with both GFP and mCherry expression. (e) HSPCs were targeted at the HBB and AAVS1 loci with a GFP and BFP expression cassette, respectively. Representative FACS plot (left panel) shows analysis seven days after targeting. All four gated populations were sorted and genomic DNA was subject to TIDE analysis for determining INDEL frequencies at the two loci (middle panel), and subject to ddPCR quantification of one of the two possible monocentric translocations between HBB and AAVS1 (right panel) (see also Figure 3—figure supplement 2). (f) Representative FACS plots from cells targeted at the HBB and AAVS1 loci with a GFP and BFP expression cassette, respectively. Representative FACS plot shows analysis four days after targeting at which point the four populations were sorted and transplanted intrafemorally into NSG mice that were irradiated 24 hr before transplantation. (g) Bone marrow from the injected femurs from the mice transplanted as described in (f) was analyzed 12 weeks after transplantation. Representative FACS plots are from a mouse from each of the four groups depicted in (f) as well as a mouse transplanted with mock-electroporated cells. The middle row depicts human engraftment gated as positive for the human leukocyte antigen complex (HLA-ABC). The upper and lower rows depict FACS plots gated from the human populations and show myeloid (CD33+) and lymphoid (CD19+) engraftment (upper row) as well as reporter gene expression (lower row) (see also Figure 3—figure supplement 3 for all transplantation data).
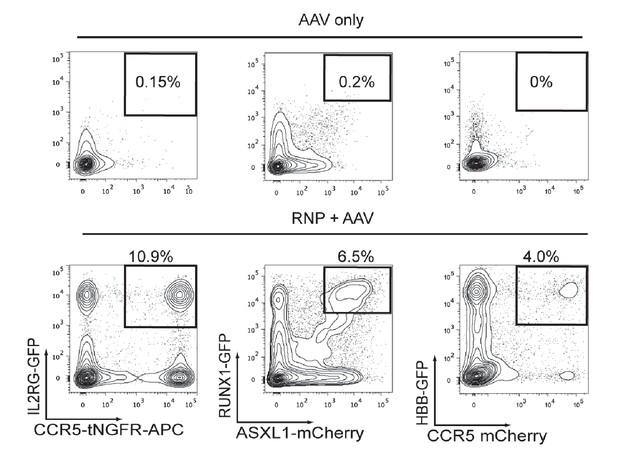
Cas9 and rAAV6-mediated di-genic homologous recombination (HR) in human CD34+ HSPCs.
Top, Representative FACS plots of HSPCs transduced with two rAAV6 donors targeting two genes with two distinct fluorescent reporters (listed in FACS plots in lower panel) show low episomal expression and few dual reporterhigh-expressing HSPCs. Bottom, HSPCs were electroporated with two different gene-specific Cas9 RNPs and then transduced with homologous rAAV6 donors (each gene targeted with a different fluorescent reporter). Representative FACS plots from Day 4 post electroporation show the generation of dual reporterhigh positive HSPCs targeted at both genes.
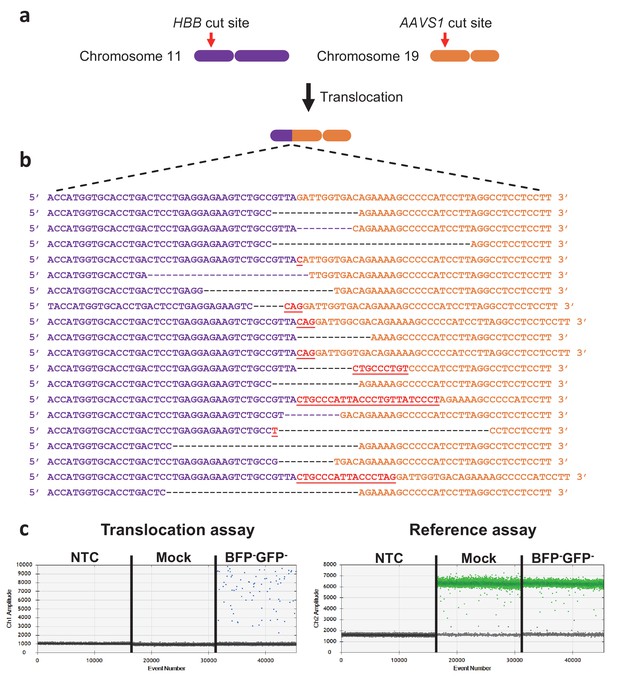
Measuring translocations after HBB and AAVS1 di-genic targeting.
(a) Schematic showing the HBB gene on chromosome 11 and the AAVS1 locus on chromosome 19. The Cas9 cut sites are shown in red. One of the two possible monocentric translocations is shown. (b) The reference sequence of the HBB-AAVS1 translocation is shown in the top. Below are representative translocation sequences from GFP-BFP- HSPCs sorted seven days after targeting (see Figure 3e, left panel). (c) Representative ddPCR analyses quantifying translocations in NTC (non-template control), mock-electroporated, and GFP-BFP- cells (see Figure 3e, right panel). The reference assay quantifies TERT gene copies used to normalize for DNA input. The translocation assay probe binds 50 bp away from the junction and none of the identified translocations would therefore exclude probe binding.
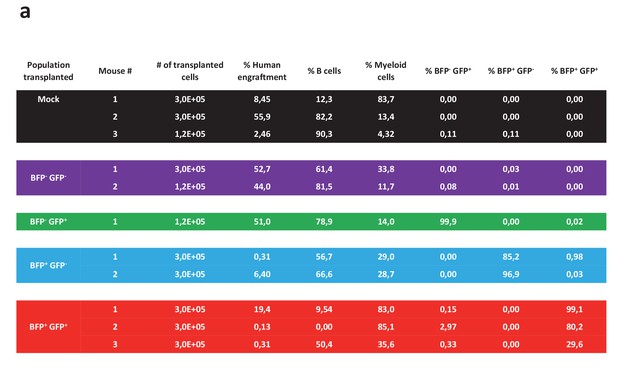
Analysis of mice transplanted with different sorted populations of cells targeted at the HBB and AAVS1 locus.
The table shows an overview of the 11 NSG mice that were transplanted intrafemorally with either mock-electroporated cells or sorted cells from the four populations displayed in Figure 3f. 12 weeks after transplant, the transplanted femurs were flushed and the cells analyzed for human engraftment based on HLA-ABC expression, B cell or myeloid phenotype (CD19 and CD33, respectively), and expression of the two reporter genes.
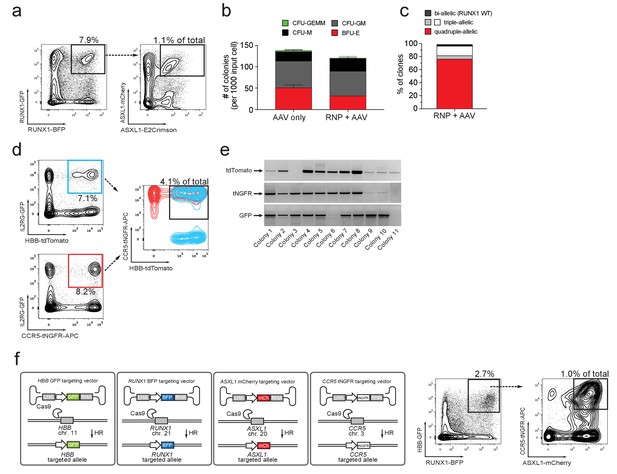
Multiplexing homologous recombination in CD34+ human hematopoietic stem and progenitor cells (HSPCs).
(a) HSPCs were electroporated with Cas9 RNP targeting ASXL1 and RUNX1 followed by rAAV6 transduction with two donors for ASXL1 (mCherry and GFP) and two donors for RUNX1 (E2Crimson and BFP). Tetra-allelically targeted HSPCs were identified as mCherryhigh/GFPhigh/BFPhigh/E2Crimsonhigh (N = 3 see Supplementary file 1e) (b) Cells modified at both alleles for RUNX1 and ASXL1 (as in (a)) were subjected to a methylcellulose assay (triplicates) and scored as BFU-E, CFU-M, CFU-GM or CFU-GEMM based on morphology 14 days after sorting. (c) PCR was performed on colony-derived gDNA to detect targeted integrations at both genes. 73 individual colonies were analyzed. Color coding for colonies with triple-allelic integration are as follows: grey: RUNX1 biallelic/ASXL monoallelic; white: RUNX1 monoallelic/ASXL1 biallelic. (d) For tri-genic targeting of HSPCs, cells were electroporated with Cas9 RNP targeting IL2RG, HBB, and CCR5 followed by transduction of three rAAV6 donors homologous to each of the three genes (IL2RG-GFP, HBB-tdTomato, and CCR5-tNGFR). Tri-genic-targeted cells were identified as reporterhigh for all three reporters (N = 5 see Supplementary file 1e). (e) Methylcellulose clones from the triple-positive cells in (d) were subjected to genotyping PCR and gel images show colonies with targeted integration at all three genes in 9/11 colonies (note that GFP shows a faint band in colony 6). (f) Left, Schematic showing strategy for targeting four different genes (HBB, RUNX1, ASXL1, and CCR5) simultaneously (tetra-genic). Four different genes are targeted by electroporation of four different Cas9 RNPs followed by transduction with four different rAAV6 donors that each targets a gene with a different reporter. Right, Tetra-genic targeting at the above-mentioned four genes was identified as reporterhigh for all four reporters (N = 3 see Supplementary file 1e).
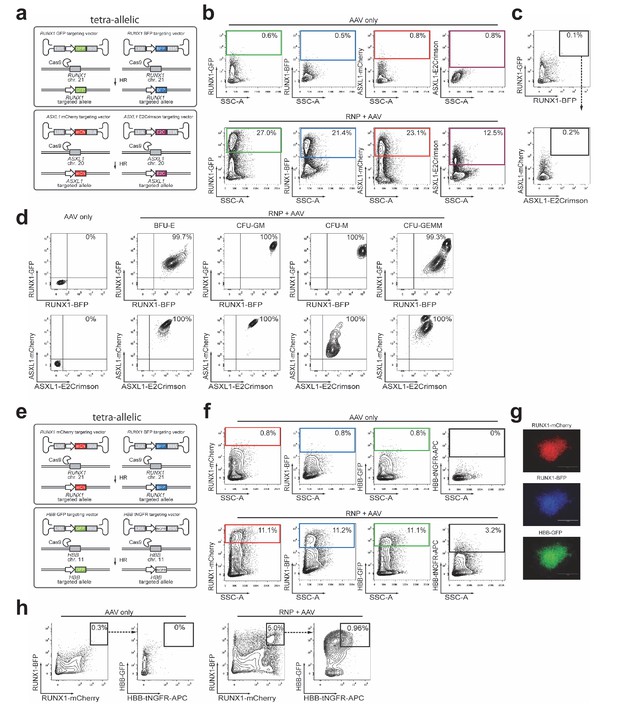
Targeting two genes for biallelic homologous recombination (HR) in primary CD34+ HSPCs.
(a) Schematic showing experimental strategy for Figure 4a for targeting both alleles of RUNX1 and ASXL1. (b) FACS plots, gating scheme, and frequencies of HR at each allele for the experiment shown in Figure 4a. (c) FACS plot showing very low frequency of tetra-reporterhigh cells without Cas9. (d) FACS plots of cells from single methylcellulose colonies derived from tetra-reporterhigh cells from Figure 4a. (e) Schematic showing targeting both alleles of RUNX1 and HBB for HR with four distinct reporters. (f) Top, FACS plots of HSPCs transduced with four rAAV6s (no Cas9 RNPs) showing the gating scheme and low episomal reporter expression without a nuclease. Bottom, HSPCs were electroporated with RNPs targeting HBB and RUNX1 and then transduced with four rAAV6s. FACS plots from day four post electroporation show MFI shift for each reporter alone. HBB-tNGFR rAAV6 has reproducibly shown lower episomal expression than all other rAAV6 we have used. (g) Images from fluorescence microscopy showing an mCherry/BFP/GFP positive CFU-GM clone that has undergone tetra-allelic HR. The colony was not stained for HBB-tNGFR. (h) Left, FACS plots show very low frequency of tetra-reporterhigh cells without Cas9. Right, Nuclease addition increases the frequency of bi and tetra-reporterhigh HSPCs.
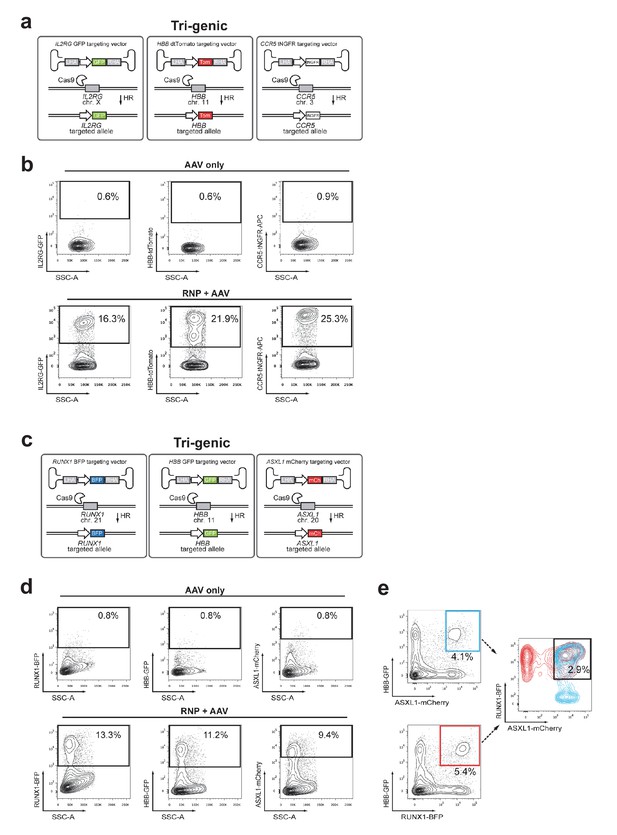
Multiplexing homologous recombination at three genes simultaneously in HSPCs.
(a) Schematic showing experimental strategy for Figure 4d targeting three genes, IL2RG, CCR5, and HBB. (b) FACS plots show gating scheme and HR frequencies at each locus for the experiment shown in Figure 4d. (c) Schematic outlining another tri-genic targeting experiment for RUNX1, ASXL1, and HBB. (d) Top, FACS plots of HSPCs transduced with three rAAV6 donors (no RNPs). Bottom, HSPCs were electroporated with gene-specific RNPs and then transduced with three rAAV6 donors. FACS plots at Day 4-post electroporation show MFI shift for each reporter alone. (e) FACS plots from same sample as in (d), but showing different combinations of di-genic reporterhigh populations that contain the same frequency of tri-genic reporterhigh cells.
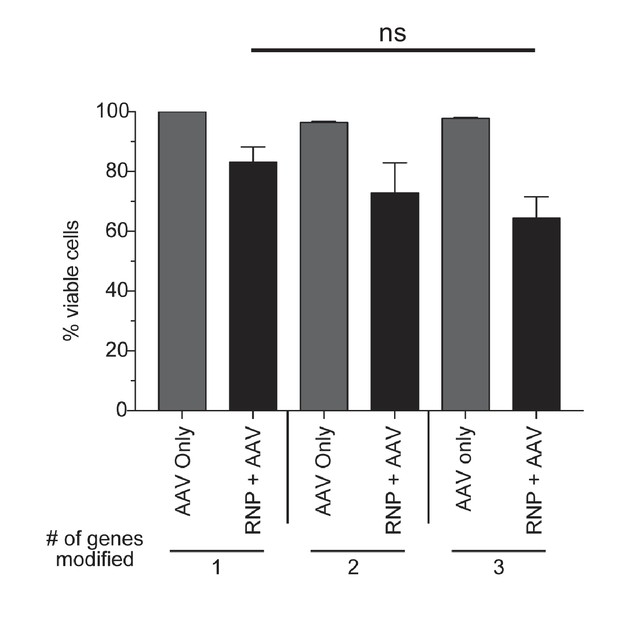
Toxicity assessment of multiplexed HR.
CD34+ cells from mobilized peripheral blood were targeted at one, two, or three genes with Cas9 RNP and rAAV6 donors. Viabilities were measured by flow cytometry 72 hr post-electroporation using Live/Dead and Annexin V stains. Viable cells are defined as live, non-apoptotic (Annexin V−) and plotted as percentage of a single AAV6 donor alone. Error bars represent SD, ns = not statistically significant, Mann-Whitney test, N = 2 different HSPC donors.
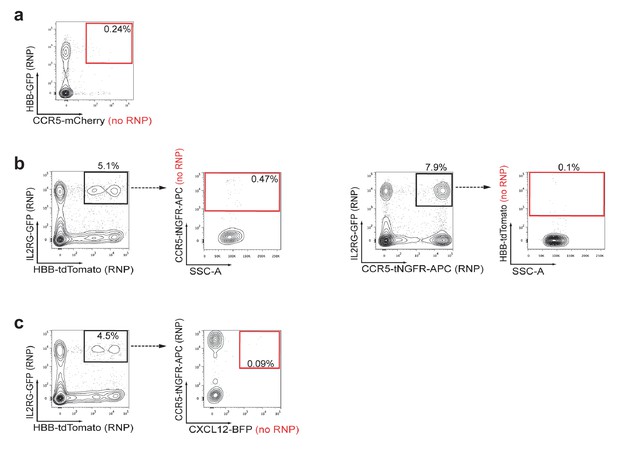
Assessment of false-positive frequencies of FACS-based identification of multiplexed HR in HSPCs.
Since capture of rAAV6 donors at the site of a DSB via NHEJ has been reported, we measured the false-positive rate of multiplexing HR via flow cytometry. (a) False-positive frequencies of di-genic targeting in HSPCs was determined by electroporating cells with an HBB-targeting Cas9 RNP followed by transduction with HBB-GFP (homologous) and CCR5-mCherry (non-homologous) rAAV6 donors. FACS plots show a false-positive rate of 0.24% dual reporterhigh cells. Note that 4% dual reporterhigh cells was reported in Figure 3—figure supplement 1 when performing di-genic targeting at CCR5 and HBB, giving a false positive rate of 6% of targeting. (b) Left, To determine false-positive frequencies of tri-genic targeting in HSPCs, we electroporated IL2RG-RNP and HBB-RNP into HSPCs followed by transduction with the rAAV6 donors IL2RG-GFP (homologous), HBB-tdTomato (homologous), and CCR5-tNGFR (non-homologous). FACS plots show a false-positive frequency of 0.47%. Note that Figure 4d shows a tri-genic targeting frequency of 4.1% (a false-positive rate of 11% of targeting). Right, We employed a similar strategy to determine false-positive frequencies of tri-genic targeting, but this time used different combinations of on-target nucleases. The false-positive rate detected here was 0.1% (2.4% of targeting). (c) To determine tetra-genic false-positive frequencies, we electroporated HSPCs with three on-target nucleases (IL2RG, HBB, and CCR5) and then transduced with three homologous rAAV6 donors (IL2RG, HBB, and CCR5) and one non-homologous donor (CXCL12). FACS plots show a frequency of 0.09% that are reporterhigh for all four reporters with Figure 4f showing a tetra-genic targeting frequency of 1.0% (a false-positive rate of 9% of targeting).
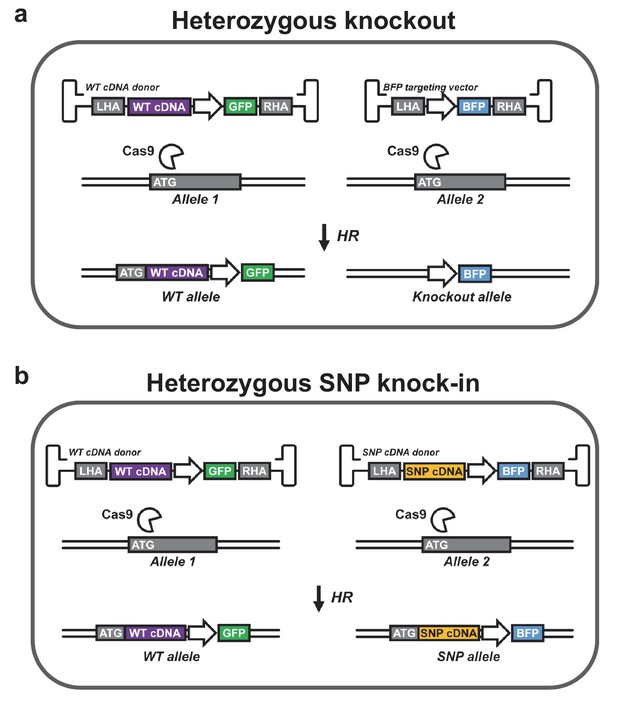
Controlling genotype with cDNA knock-in.
(a) A heterozygous knockout population can be generated with two HR donors. The first donor is designed to knock-in a wild-type (WT) cDNA cassette into the start codon (ATG) of the gene of interest followed by a cassette encoding a reporter gene (here GFP). WT cDNA is expressed from the endogenous promoter as reported by Voit et al. (2014), Hubbard et al. (2016), and Dever et al. (2016), which maintains endogenous regulatory control over gene expression. The other donor encodes another reporter (here BFP), which disrupts the targeted gene. Double positive cells (GFP+/BFP+) are heterozygous for the knockout allele. (b) A population heterozygous for a particular SNP can be generated using two donors that knock in cDNA expression cassettes followed by different reporter genes. One cDNA is WT while the other carries the SNP of interest. Double positive cells (GFP+/BFP+) are heterozygous for the SNP allele. Endogenous 3’ UTRs may be incorporated to preserve posttranscriptional regulation. Heterozygous SNP cDNA knock-in may be expanded to two or more genes, which may be of particular interest in studies of leukemia-mutated genes such as DNMT3A, IDH1/2, JAK2, and KRAS, which often occur in various combinations as heterozygous gain-of-function or dominant negative mutations. In addition, reporter knock-in combined with WT cDNA knock-in (as depicted in a) as well as SNP cDNA knock-in (SNP that disrupts gene or gene function) combined with WT cDNA knock-in (as depicted in b) could be used to study haploinsufficiencies. Though not depicted, all genes in the schematic are followed by polyadenylation signals.
Tables
Overview of targeting experiments in hematopoietic stem and progenitor cells (HSPCs).
Overview of all HSPC targeting experiments performed in this study with the number of independent experiments (N) for each experiment type, and the mean targeting efficiency (±SD). See also Supplementary file 1a, c, and e.
| Experiment | N | % efficiency ± SD |
|---|---|---|
| Monogenic | 47 | 21.7 ± 13.4 |
| Biallelic | 16 | 5.5 ± 4.2 |
| Di-genic | 17 | 8.1 ± 8.1 |
| Tetra-allelic | 3 | 0.9 ± 0.3 |
| Tri-genic | 6 | 4.5 ± 4.8 |
| Tetra-genic | 3 | 0.7 ± 0.3 |
Additional files
-
Supplementary file 1
(a) Overview of Cas9 and rAAV6 mono-genic targeting experiments performed in cord blood (CB), bone marrow (BM), and mobilized peripheral blood (mPB)-derived human CD34+HSPCs.
This table summarizes all independent experiments targeting HBB, CCR5, IL2RG, RUNX1, ASXL1, STAG2, and AAVS1 in HSPCs and the reporter genes used. GFP: green fluorescent protein, tNGFR: truncated Nerve Growth Factor Receptor, BFP: blue fluorescent protein. Efficiencies were averaged across 47 independent experiments, N = 47. (b) Overview of genotypes for the non-integrated alleles in mono-genic integration experiments. The three tables show the different INDELs that were identified by Sanger Sequencing of the non-edited allele in mono-genic targeting experiments (CCR5, IL2RG, and RUNX1) used to analyze genotype frequencies shown in Figure 1—figure supplement 2b and d. Alleles are grouped into WT (blue), INDELs that preserve the reading frame (red) and INDELs that disrupt the reading frame (green). Note that INDELs that preserve the reading frame can potentially be disruptive depending on the size and location. For example, the 147 bp deletion in RUNX1 is considered disruptive because of its large size and because it deletes the splice donor site in the intron between exon 2 and 3. For IL2RG, one clone was found to have an allele with integration of 230 bp from the donor (at the end of the RHA and 72 bp into the ITR). (c) Overview of di-genic and biallelic targeting experiments in cord blood (CB), bone marrow (BM), and mobilized peripheral blood (mPB)-derived human CD34+HSPCs. This table summarizes the experiments targeting HSPCs for biallelic and di-genic HR and the reporter genes used. GFP: green fluorescent protein, tNGFR: truncated Nerve Growth Factor Receptor, BFP: blue fluorescent protein. Efficiencies were averaged across 16 and 17 independent experiments, respectively, N = 16 and N = 17. (d) Overview of genotypes for the non-integrated alleles in clones with tri-genic integrations. Each row of the table represents the genotype of a colony established from a tri-genic targeting experiment (IL2RG, HBB, and CCR5). Alleles are grouped into WT (blue), INDELs that preserve the reading frame (red) and INDELs that disrupt the reading frame (green). Note that INDELs that preserve the reading frame can potentially be disruptive depending on the size and location. For HBB we identified one clone where HBD had been used as repair template and three clones with mono-allelic integration of part of the SFFV promoter indicative of HR events that ended prematurely. (e) Overview of tetra-allelic, tri-genic, and tetra-genic targeting experiments performed in human CD34+HSPCs derived from cord blood (CB), bone marrow (BM), and mobilized peripheral blood (mPB). This table summarizes the independent multiplexing HR experiments performed for tetra-allelic, tri-genic, and tetra-genic targeting and the reporter genes used. GFP: green fluorescent protein, tNGFR: truncated Nerve Growth Factor Receptor, BFP: blue fluorescent protein. Efficiencies were averaged across independent experiments, N = 3 (tetra-allelic and tetra-genic) and N = 6 (tri-genic).
- https://doi.org/10.7554/eLife.27873.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.27873.021





