In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing
Figures
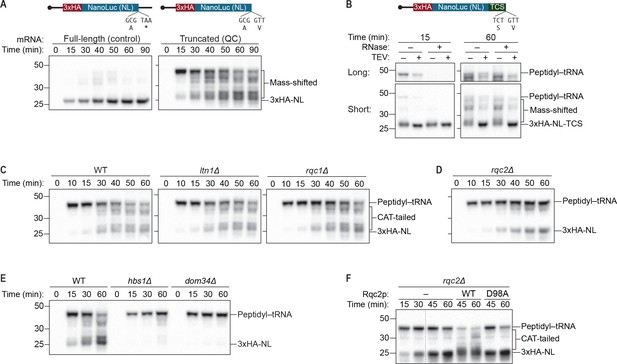
An S. cerevisiae in vitro translation system recapitulates synthesis of Rqc2p-dependent polypeptide extensions.
(A) Time courses of S. cerevisiae in vitro translation (ScIVT) reactions. ScIVT reactions were prepared using wild-type (WT) extracts and 1 μg of either a full-length (left; includes a stop codon and 3′-UTR) or truncated (right; encodes a terminal valine residue) mRNA encoding lysine-free 3xHA-NanoLuc (3xHA-NL). At the indicated time points, aliquots of the reactions were quenched in 2X Laemmli Sample Buffer. Proteins were separated by SDS-PAGE, and HA-tagged translation products were visualized by immunoblotting. (B) Analyses of mass-shifted products. An ScIVT reaction was prepared using WT extracts and a lysine-free truncated mRNA substrate that also encodes a TEV cleavage site (TCS). Translation was halted after 15 or 60 min by addition of 20 mM EDTA, after which reactions were treated without (–) or with (+) TEV and/or RNase A/T1 cocktail for 60 min. Translation products were analyzed by immunoblotting as in (A). ‘Long’ and ‘Short’ refer to exposure times of the blots. (C–E) Genetic analysis of mass-shifted products. ScIVT reactions were prepared using extracts from strains of the indicated genotypes and a lysine-free truncated mRNA substrate. Reactions were performed and analyzed as in (A) but with less mRNA (480 ng). The species that migrate just below the peptidyl–tRNA in rqc2Δ extracts in (D) represent peptidyl–tRNA degradation products that arise due to prolonged incubation in the absence of Rqc2p. (F) Rescuing Rqc2p deficiency in vitro. ScIVT reactions were prepared using rqc2Δ extracts and a lysine-free truncated mRNA substrate. After 30 min of translation, reactions were supplemented with either protein storage buffer (–) or purified Rqc2p (WT or CAT-tailing deficient D98A at 420 nM final concentration) and indicated time points were analyzed by SDS-PAGE and immunoblotting. Dashed lines indicate where intervening lanes were removed for clarity.

Purified wild-type and mutant Rqc2p.
C-terminal polyhistidine-tagged Rqc2p (WT and D98A mutant) were purified as described in Materials and methods and analyzed by SDS-PAGE and Coomassie staining. 1/10th volume of the corresponding purified protein was added to ScIVT reactions.
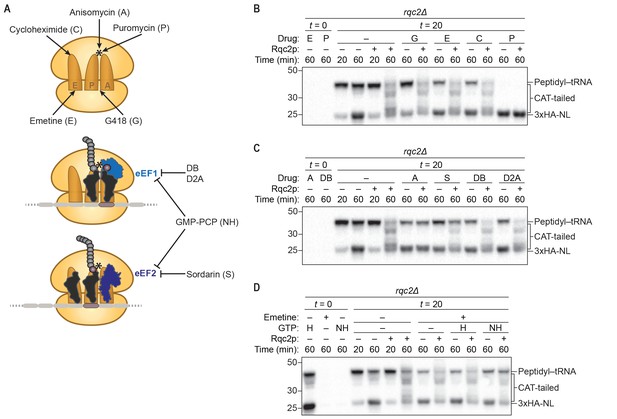
CAT-tail synthesis is mechanistically distinct from canonical translation.
(A) Schematics of small-molecule inhibitors that directly bind the ribosome (top) or that target the translation elongation factors eEF1a or eEF2 (bottom). Inhibitors: (A) anisomycin; (C) cycloheximide; (D2A) didemnin 2A; (DB) didemnin B; (E) emetine; (G) G418; (H) hydrolyzable GTP; (NH) non-hydrolyzable GTP-analog GMP-PCP; (P) puromycin; (S) sordarin. (*) Denotes the peptidyl-transferase center of the 60S subunit. (B–D) Effects of small-molecule inhibitors on CAT tailing. ScIVT reactions were prepared using rqc2Δ extracts and a lysine-free truncated mRNA substrate. After 0 min (t = 0) or 20 min (t = 20) of translation, reactions were supplemented with either protein storage buffer (–) or purified Rqc2p at 670 nM final concentration (+) and the indicated inhibitor(s). Indicated time points (‘Time (min)') were analyzed by SDS-PAGE and immunoblotting. Additional t = 0 controls for the remaining inhibitors can be found in Figure 2—figure supplement 1.
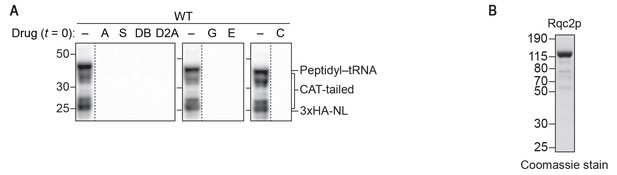
Inhibitors and purified Rqc2p used to dissect the mechanism of CAT-tail synthesis.
(A) Effects of small-molecule inhibitors on translation. ScIVT reactions were prepared using WT extracts and a truncated mRNA substrate, supplemented without (–) or with (+) the indicated inhibitors after 0 min (t = 0) of translation, and analyzed by SDS-PAGE and immunoblotting. Inhibitors: (A) anisomycin; (C) cycloheximide; (D2A) didemnin 2A; (DB) didemnin B; (E) emetine; (G) G418; (S) sordarin. Dashed lines indicate where intervening lanes were removed for clarity. (B) C-terminal polyhistidine-tagged Rqc2p was purified as described in Materials and methods and analyzed by SDS-PAGE and Coomassie staining. 1/10th volume of purified protein was added to ScIVT reactions. Note that this stock of protein was expressed and purified independently from the stock in Figure 1—figure supplement 1.
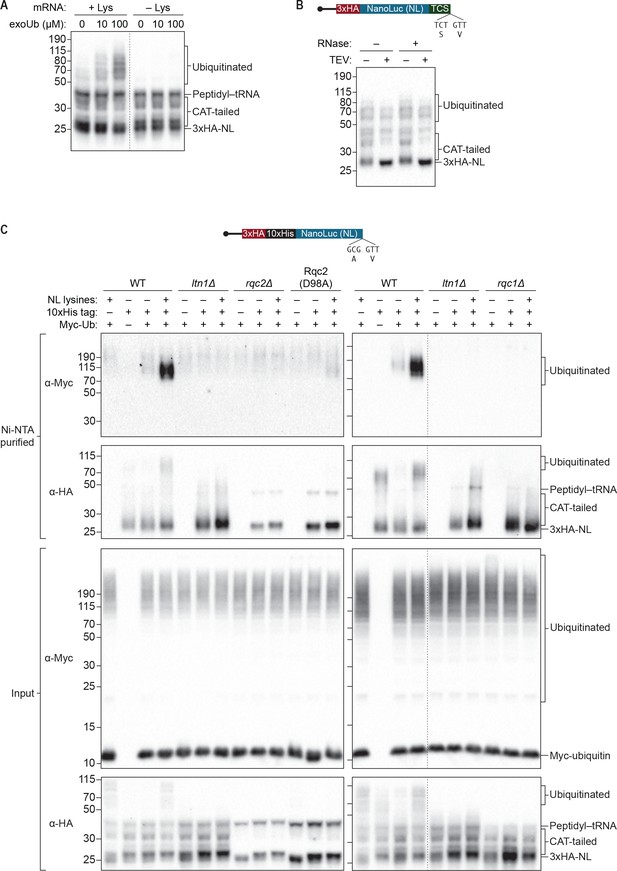
S. cerevisiae in vitro translation recapitulates Ltn1p-mediated ubiquitination.
(A) Effects of adding exogenous ubiquitin to ScIVT reactions. ScIVT reactions conducted in WT extracts with lysine-containing (+Lys) or lysine-free (–Lys) truncated mRNA were supplemented with the indicated concentrations of recombinant ubiquitin, incubated for 60 min, and then analyzed by SDS-PAGE and immunoblotting. Dashed line indicates where intervening lanes were removed for clarity. (B) Analysis of high-molecular-weight smears. RNase A/T1 and TEV protease treatment of ScIVT reactions programmed with lysine-containing truncated mRNA encoding a TEV cleavage site (TCS) in WT extracts supplemented with 100 μM recombinant ubiquitin. Translation was halted after 60 min by addition of 20 mM EDTA, after which reactions were treated without (–) or with (+) TEV and/or RNase A/T1 for 60 min, and then analyzed by SDS-PAGE and immunoblotting. Note that due to long incubations (120 mins), very little peptidyl–tRNA persists in these reactions. (C) Isolation and detection of ubiquitinated ScIVT products. ScIVT reactions were conducted with 1.2 μg of truncated mRNA (3xHA-10xHis-NanoLuc, with or without lysines or His tag as indicated) in extracts prepared from strains of the indicated genotypes and supplemented with 10 μM recombinant Myc-ubiquitin. For input samples (bottom panels), one-third of the ScIVT reaction was quenched with 2X Laemmli Sample Buffer. For Ni-NTA-purified samples (top panels), two-thirds of the ScIVT reaction was quenched with 6 M guanidine-HCl. For SDS-PAGE, 30% of input samples and 100% of Ni-NTA-purified samples were separated on 12% NuPAGE gels and translation products were visualized by immunoblotting with antibodies indicated at left. Dashed lines indicate where intervening lanes were removed for clarity.
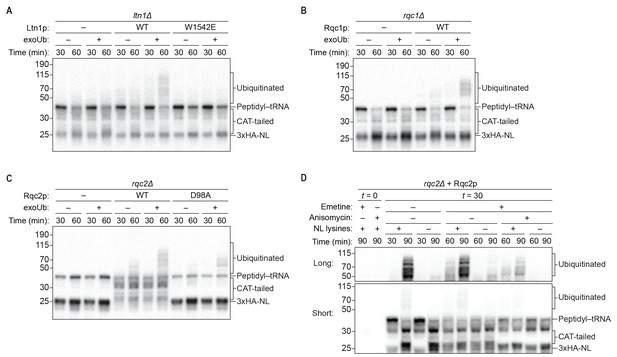
Rqc1p and CAT tailing contribute to Ltn1p-dependent ubiquitination.
(A–C) Genetic analysis of RQC-mediated ubiquitination in ScIVT. ScIVT reactions were prepared using extracts from strains of the indicated genotype, a lysine-containing truncated mRNA substrate, ubiquitin storage buffer (–) or 100 μM recombinant ubiquitin (+), and either protein storage buffer (–) or the indicated purified proteins (+): Ltn1p at 130 nM, Rqc1p at 70 nM, and Rqc2p at 420 nM final concentration. (D) ScIVT reactions were conducted using rqc2Δ extracts, a lysine-free or lysine-containing truncated mRNA substrate, and 100 μM exogenous ubiquitin. After 0 min (t = 0) or 30 min (t = 30) of translation, all reactions were supplemented with an equal volume of ‘mock ScIVT’ (i.e., without mRNA) containing 1.34 μM purified Rqc2p, 100 μM exogenous ubiquitin, and the indicated inhibitor(s). Indicated time points (‘Time (min)') were analyzed by SDS-PAGE and immunoblotting. ‘Long’ and ‘Short’ refer to exposure times of the blots.
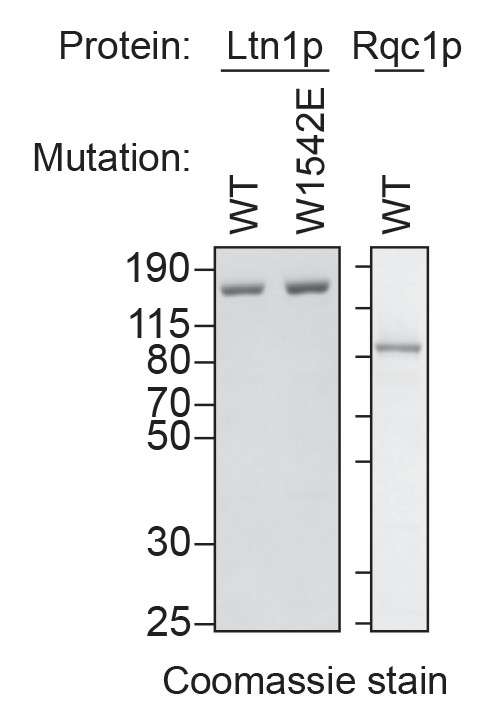
Purified Ltn1p and Rqc1p.
C-terminal polyhistidine-tagged Ltn1p (WT and W1542E mutant) and Rqc1p were purified as described in Materials and methods and analyzed by SDS-PAGE and Coomassie staining. 1/10th volume of the corresponding purified protein was added to ScIVT reactions.
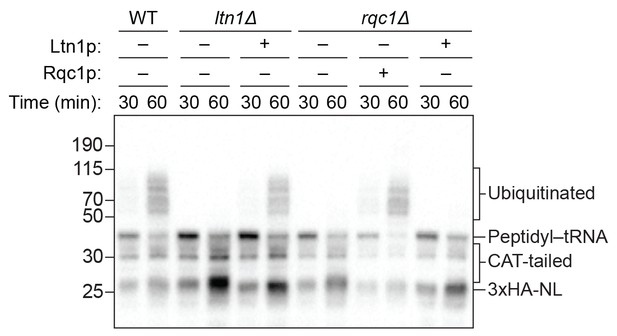
Impact of excess Ltn1p on ubiquitination in rqc1Δ extracts.
ScIVT reactions were conducted as in Figure 4A–C except that 100 μM exogenous ubiquitin was present in all reactions.
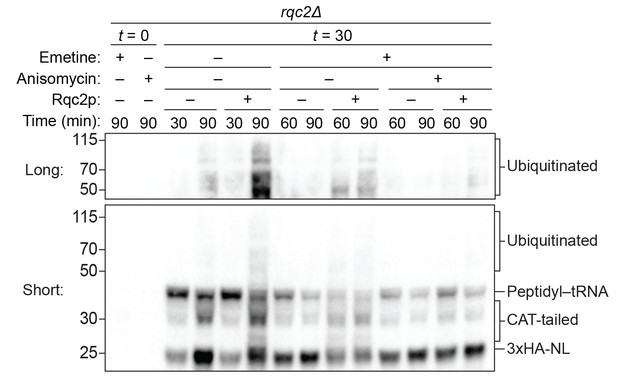
Impact of CAT-tailing inhibition on ubiquitination.
ScIVT reactions were conducted as in Figure 4D, except all reactions were performed with lysine-containing mRNA and ‘mock ScIVT’ containing either protein storage buffer (–) or 1.34 μM purified Rqc2p (+).
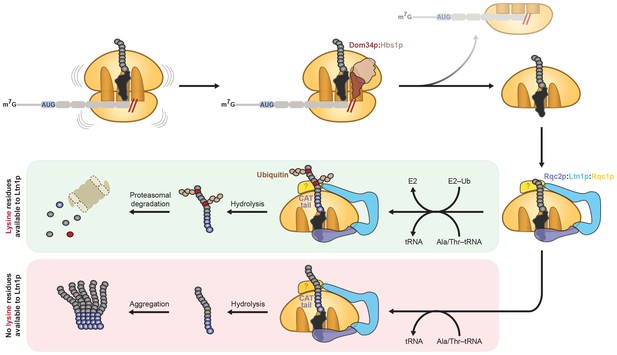
Model for CAT tailing and ubiquitination of stalled nascent chains.
When an 80S ribosome stalls during translation, splitting factors recognize the stalled translation complex to facilitate dissociation of the 40S subunit and mRNA. Ltn1p, Rqc2p, and Rqc1p (unknown location indicated by ‘?') bind the resulting 60S:peptidyl–tRNA complex. Together with the peptidyl-transferase center of the 60S subunit, Rqc2p facilitates elongation of the stalled nascent chain with a CAT tail by recruiting alanine- and threonine-charged tRNAs to the A site. If the nascent chain contains a lysine residue (red circle) located within the vicinity of the Ltn1p RING domain (or potentially hidden inside the ribosome exit tunnel), CAT tailing and Rqc1p enhance or facilitate Ltn1p-mediated ubiquitination of the nascent chain, respectively, for subsequent proteasomal degradation (green box). If the nascent chain does not contain any lysine residues (or contains lysine residues that are too distant from the Ltn1p RING domain), CAT tails may promote aggregation of incompletely synthesized proteins (red box). In both instances, Rqc2p activity promotes hydrolysis of the peptidyl–tRNA linkage and liberation from the 60S subunit.
Additional files
-
Supplementary file 1
S. cerevisiae strains used in this study.
The names, genetic backgrounds, and protein expression plasmids of the strains generated and used in this study are listed in this table.
- https://doi.org/10.7554/eLife.27949.013
-
Supplementary file 2
DNA sequences of the constructs used in this study.
- https://doi.org/10.7554/eLife.27949.014





