Deconstruction of the beaten Path-Sidestep interaction network provides insights into neuromuscular system development
Figures
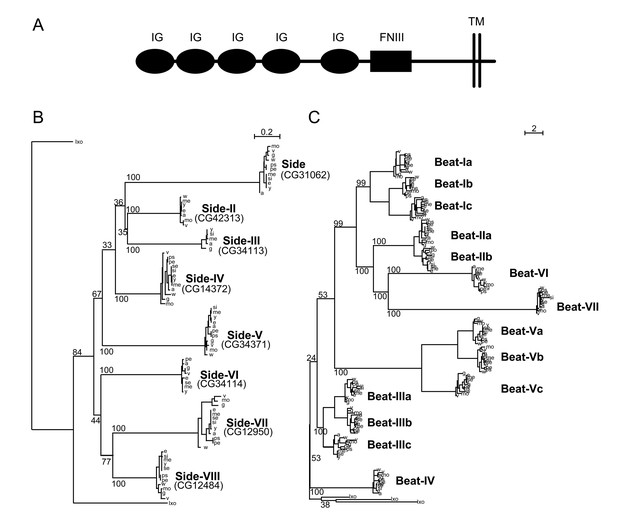
Phylogenetic analysis of Beaten Path and Sidestep paralogs.
(A) Phylogeny of the Beat family of receptors rooted against the tick Ixoides scapularis (Ixo) Beats. Beat-VII and Beat-VI share a more recent ancestor than previously described. (B) Extracellular architecture of the Side subfamily. Detailed ClustalW alignment of individual domains and conservation are in Figure 1—figure supplement 1. (IG, immunoglobulin superfamily; FNIII, Fibronectin type III; TM, transmembrane domain). (C) Phylogeny of the Side family of related proteins rooted against similar IgSF proteins predicted in the tick, Ixoides scapularis (Ixo) that form a distinct outgroup. Names are assigned to the paralogs on the basis of their evolutionary distance from Sidestep and their CG Flybase identifiers are in parentheses.

Sequence alignments of extracellular domains in the Side family.
Alignment of the five IgSF domains (Ig) and the fibronectin type III domain (FN III) for the different Side family members. Alignments were generated with ClustalOmega with the individual IgSF or FNIII domains of Side, Side-II, III, IV, V, VI, VII and VIII (GenBank accession numbers: AAF56708.2, AAX52666.3, ABI31143.1, AAF54952.3, AAF46967.2, ABI31162.1, AGB95814.1 and AAF57468.4 respectively). Domain boundaries are predicted by an initial search with SMART and extended by homology modeling using PHYRE2 and secondary structure predictions using PSIPRED. Positions conserved in the IgSF or FNIII fold are shadowed in black if they present a very high degree of conservation in the fold (residues corresponding to positions B3, C5 and F3 in the IgSF domains and C1 and F1 in the FNIII domains respectively) or grey if they present a high degree of conservation in the fold (residues corresponding to positions B1, C3, E5 and F5 in the IgSF domains and C3, C5, F3 and F5 in the FNIII domains respectively).
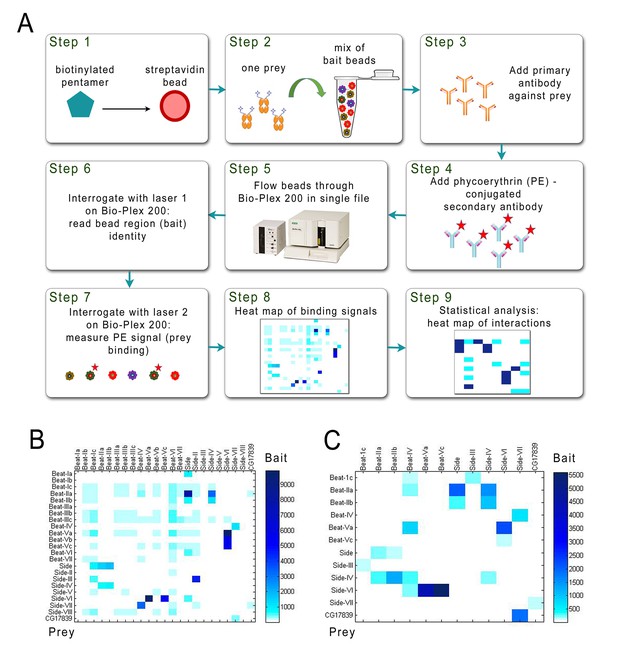
Schematic of the BPIA and heat maps of Beat/Side interactions.
(A) Each biotinylated AP bait was captured from media with streptavidin-coupled beads from a particular bead region (Step1), and all bead regions were pooled and aliquoted. A different Fc prey was added to each aliquot and incubated overnight (Step 2). Primary antibody was added to the reactions the next day (Step 3), followed by phycoerythrin (PE)-conjugated secondary antibody (Step 4). The reactions were then transferred to a 96-well plate. In the Bio-Plex 200 machine, beads are aspirated in single file from the wells (Step 5) and interrogated by two lasers. The first laser reads the color (‘region’) for each bead, and therefore identifies which bait is being analyzed (Step 6). The second laser reads the PE signal, and the strength of this signal indicates how much prey is bound to each bead region (Step 7). These data are used to produce a heat map of raw interaction signals (Step 8). After statistical analysis, likely receptor-ligand interactions are defined (Step 9). (B) 23 × 23 matrix of raw interaction signals between Beats and Sides performed using purified prey. (C) 12 × 12 matrix of raw interaction signals between Beats and Sides using unpurified prey. All interactions seen with purified prey were also detected using unpurified prey, except for Beat-Ic::Side.
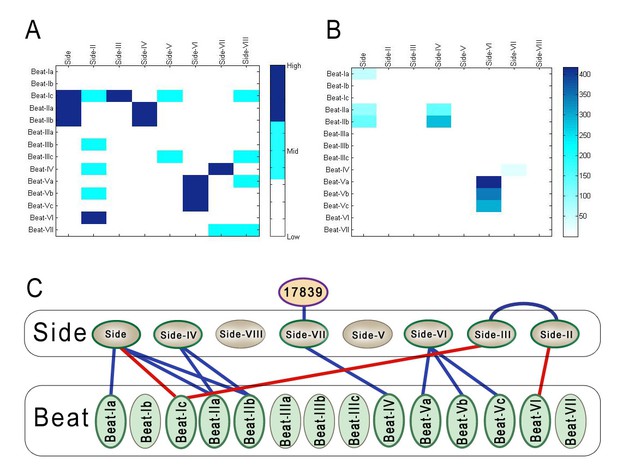
Comparison of BPIA and ECIA data, and the revised Beat-Side network map.
(A) A 14 × 8 quantized heat map of interactions between Beats and Sides determined using the BPIA. This heat map was generated using the geometric means of Z scores for interactions between Beats and Sides calculated using the numbers in Figure 2B. Each number in the matrix was then assigned to one of three values: low, mid and high. These values were calculated using cutoffs of 80% and 90%. (B) 14 × 8 heat map of geometric means of Beat/Side interactions using ECIA data from Özkan et al. (C) Updated network of Beat-Side interactions. Three new interactions were discovered with the BPIA: Beat-VI::Side-II, Beat-Ic::Side, and Beat-Ic::Side-III.
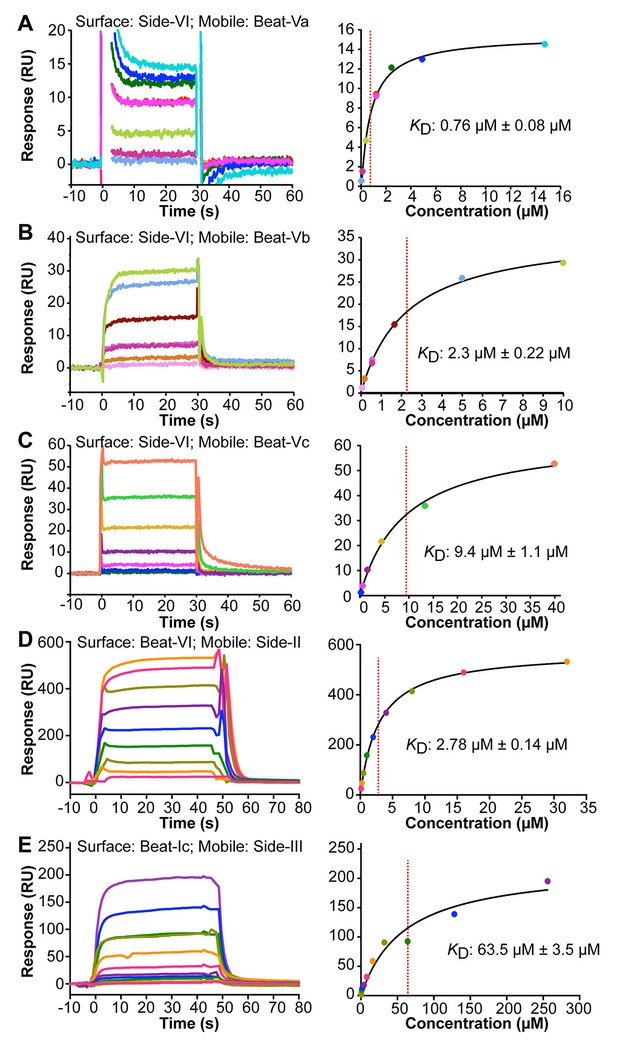
Surface Plasmon Resonance sensorgrams (left panels) and binding isotherms (right panels) for five Beat–Side complexes.
Equilibrium binding responses are fit to Langmuir isotherms to calculate dissociation constants (KD). Each color in the sensorgrams represents the concentration of the analyte in mobile phase. Zero time-point indicates time of analyte injection. The color scheme from the sensorgrams is preserved in the binding isotherms. (A–C) Interactions of Side-VI with the Beat-V family of receptors. Side-VI was captured on a Biacore SA chip, and titration series of Beat-Va (A), Beat-Vb (B), and Beat-Vc (C) were flowed over the SA chip. The ±errors represent standard error from the fitting of one titration series. (D and E) Interactions of Beat-VI with Side-II (D), and Beat-Ic with Side-III (E). Beat-VI and Beat-Ic were captured on a Biacore SA chip, and the Sides were flowed over the chip. The ±errors represent standard error of the mean for KD from three titration series.
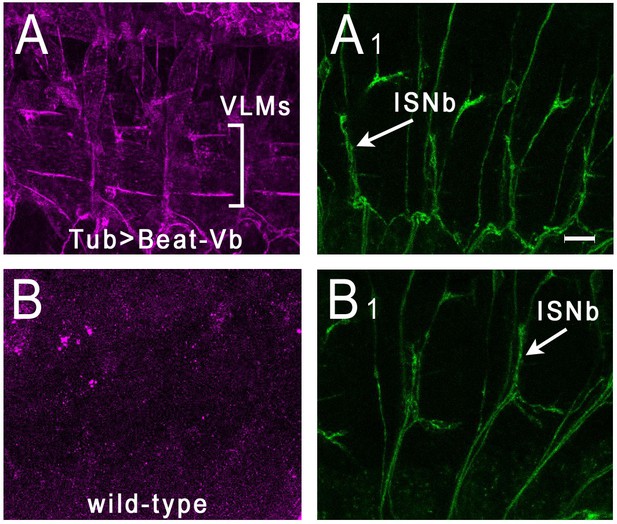
Side-VI-AP binds to Beat-Vb in live embryos.
Stage 16 embryos were live-dissected and stained with Side-VI-AP and 1D4. (A), (A1) Tub-GAL4 >UAS Beat-Vb embryos stained with Side-VI-AP (A) and mAb 1D4 (A1). Strong Side-VI-AP staining of muscle fibers is observed. The ventrolateral muscle field is indicated by brackets. Note that the lateral muscle fibers (muscles 21–24) above the VLMs are outlined by Side-VI-AP staining. (B), (B1) WT embryos stained with Side-VI-AP (B) and 1D4 (B1). There is no staining of muscle fibers above background. Scale bar, 12 µm.
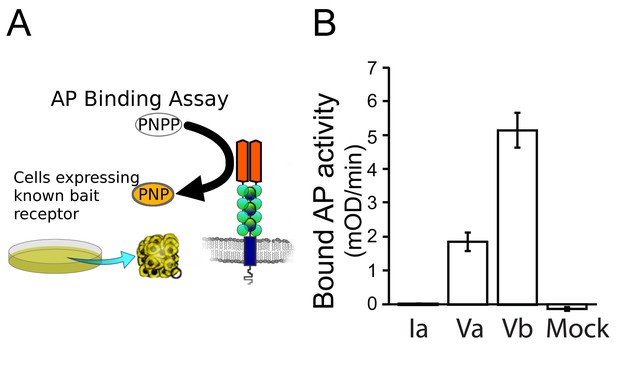
The Side-VI ectodomain binds to Beat-Va and Beat-Vb expressed on the surfaces of S2 cells.
(A) Graphical representation of the AP binding assay. AP supernatant (0.5 nM) is incubated with cells expressing a receptor on their surface and bound AP activity is detected after washing. (B) Beat-Va and Beat-Vb expressed on Drosophila S2 cells bind soluble Side-VI-AP (p<0.005; N = 3), but Beat-Ia does not.
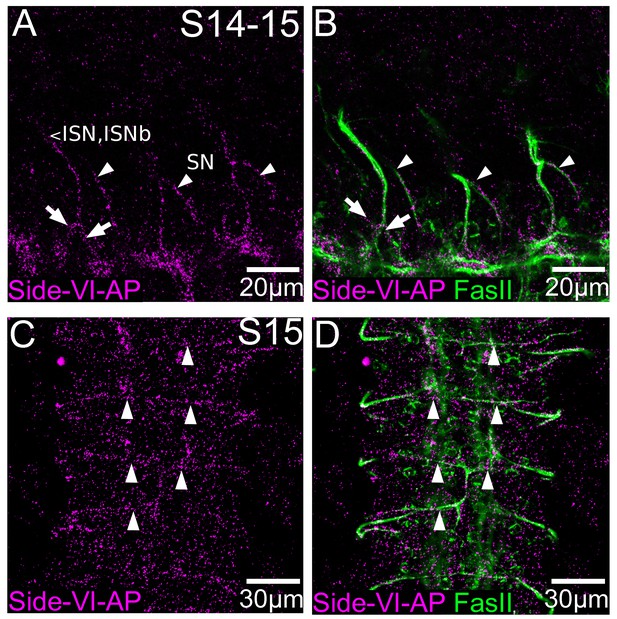
The Side-VI ectodomain binds to motor axons in live-dissected embryos.
Live-dissected stage 14–15 wild type embryos were stained with Side-VI-AP (magenta) and 1D4 (green). (A, B) The intersegmental (ISN) and segmental (SN) motor nerves are labeled. The anterior (left) and posterior (right) tributaries of the ISN are labeled with arrows, while the SN is labeled with arrowheads in each segment. (C, D) Side-VI-AP binding to the transverse nerve (TN), which originates at the CNS midline, in a stage 15 embryo. The location of the TN is indicated with arrowheads. Side-VI-AP staining (magenta) is overlaid with ID4 staining (green) to show motor nerves in (B) and (D).
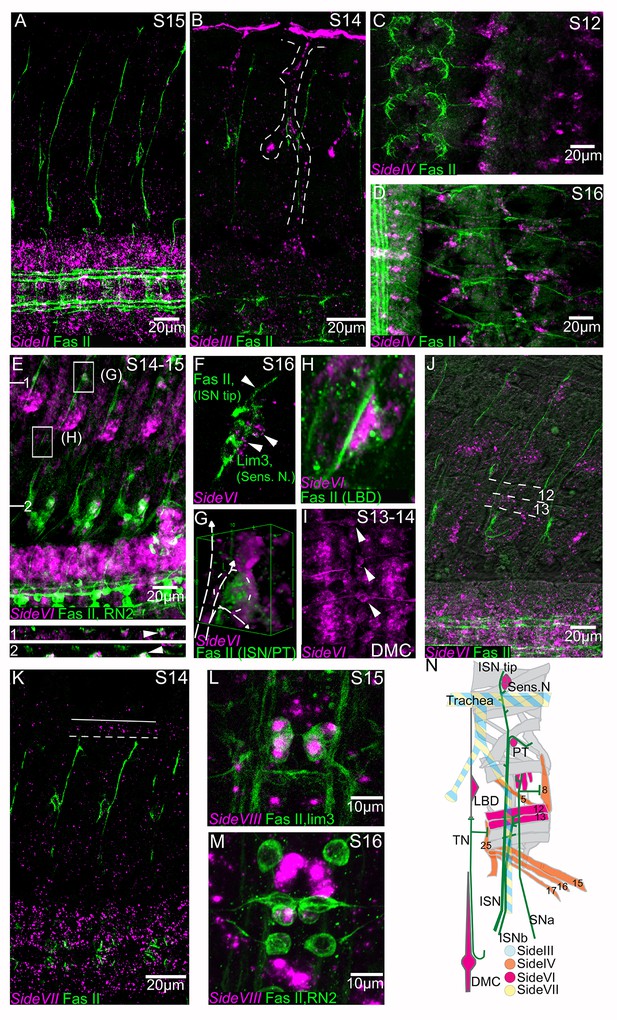
Embryonic expression patterns of side genes.
Fluorescent in situ hybridization (magenta) of side-II, side-III, side-VI, side-VII and side-VIII genes in fillet preparations. All preparations are co-stained with anti-Fasciclin II antibody (1D4) to reveal all motor nerves (green). (A) side-II is predominantly expressed in the CNS, where it has an increasingly broad expression pattern as development progresses. (B) side-III expression pattern in a stage 14 embryo in the developing trachea (dashed line). (C) side-IV expression pattern in a stage 12 embryo in ventral muscle precursors. (D) At stage 16 expression of side-IV is localized to the ventral oblique muscles (muscles 15, 16, 17), the ventral transverse (25), the lateral oblique (5) and the segment border (8) muscles. (E) Expression of side-VI at stage 14–15 co-stained for RN2-Gal4 > UAS tau-LacZ. side-VI is broadly expressed in the CNS and in specific tissues in the periphery. XZ sections are indicated and represented underneath the main panel and magnifications of selected areas (G, H) are presented in individual panels. Orthogonal views show a cross section of a dorsal set of sensory neurons (1) and the junction of the ISN at its first branch, FB (2). The location of the ISN is marked with an arrowhead. (F) In a stage 16 embryo the ISN tip explores a group of side-VI expressing dorsal sensory neurons. (G) 3D projection of the ISN FB region where side-VI is expressed at high levels in the PT cell. The path of the ISN is overlaid with a dashed line. (H) The lateral bidendritic neuron (LBD), a synaptic target of the transverse nerve (TN), expresses high levels of side-VI. (I) side-VI is expressed in the dorsal median cell (DMC, arrowheads) in the CNS. (J) side-VI expression in ventral muscles in a stage 16 embryo (dorsal and ventral borders of muscles 12 and 13 are indicated by dotted lines). (K) side-VII expression in a S14 embryo is broad in the CNS and in the trachea (outlined). (L, M) side-VIII expression at stages 15–16 in RP1, 3, 4, and five motoneurons co-stained for lim3 >Tau myc (L), and in the pCC interneuron co-stained for RN2-Gal4 > UAS tau-myc (M). (N) Synopsis of side-III, IV, VI and VII expression in the periphery. Anterior is left and the ventral CNS is down in all panels except C, D, I, L, M and N where anterior is up. Scale bars indicate distances.
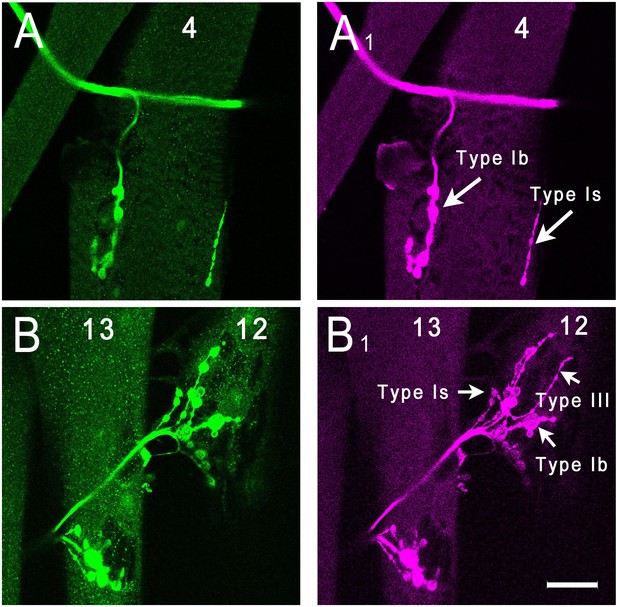
Analysis of side-VI expression patterns in larvae.
Side-VI-T2A-GAL4 > eGFP third instar larvae were dissected and stained with anti-GFP (green) and anti-HRP (magenta) to visualize neuronal membranes. GFP staining was observed on motor axons and NMJs. GFP channel (A) and HRP channel (A1) of muscle 4. Arrows denote Type Ib and Type Is arbors on muscle 4. GFP channel (B) and HRP channel (B1) of muscles 12 and 13. Arrows denote Type Is, Type Ib and Type III branches of the muscle 12 NMJ. The punctate green staining on muscles 12 and 13 may indicate that there is residual expression of side-VI in these muscles at third instar. Scale bar, 25 µm.
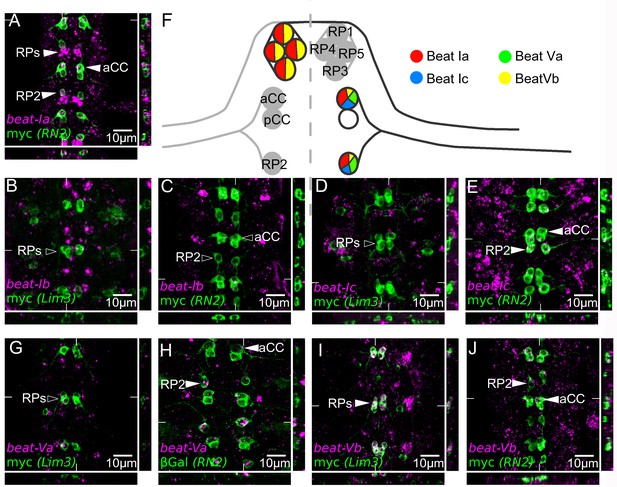
Embryonic expression of beat-I and beat-V subgroups in motor neurons indicates regulatory divergence.
Fluorescent in situ mRNA hybridization for the beat-I and beat-V groups of genes (magenta). Individual motor neurons are marked (green) with anti-myc in a Lim3A-tau-myc (Lim3) line and anti-myc or anti-βGal in RN2-Gal4 > UAS tau-myc-eGFP and RN2-Gal4 > UAS LacZ (RN2) to reveal RP1, 3, 4, and 5 (Lim3) or aCC and RP2 (RN2) cells, respectively. (A) beat-Ia is expressed in the ISN pioneer motor neurons aCC and RP2 and in the ISNb motor neurons RP1, 3, 4, and 5 (arrowheads). (B) beat-Ib is not expressed at observable levels in the RP1, 3, 4 or 5. (C) beat-Ib is expressed only at low levels (isolated magenta dots) in aCC and RP2. (D) beat-Ic is not expressed in RP1, 3, 4 and 5, but is expressed (E) in aCC and RP2. (G) beat-Va is not expressed in RP1, 3, 4, or 5 motor neurons but is expressed in aCC and RP2 (H), clearly showing higher levels in RP2 than in aCC. (I) beat-Vb is expressed at high levels in RP1, 3, 4 and 5 motor neurons and at low levels in aCC and RP2 (J). All embryos are dissected to expose the CNS. Anterior is up in all images, with the ventral midline in the center. Coordinates of orthogonal slices are indicated on main panels and XY and XZ cuts are represented to the right and bottom of each panel respectively. (F) Expression profiles of the beat-I and beat-V genes in dorsally (aCC, RP2) and ventrally (RP1, 3, 4, 5) projecting motor neurons. Scale bars indicate distances.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28111.014





