CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness
Figures

Disruption of RNA binding proteins via targeted CRISPR/Cas9-mediated homologous recombination.
(A) Schematic of template-directed homologous recombination to disrupt a gene of interest and replace with a positive-selection cassette (NeoR = Neomycin resistance) and one of two tissue specific fluorescent marker genes labeling the pharynx or body wall musculature. (B) Double mutants are created by simple crosses on the fluorescent dissection microscope looking for presence of both fluorescent markers.
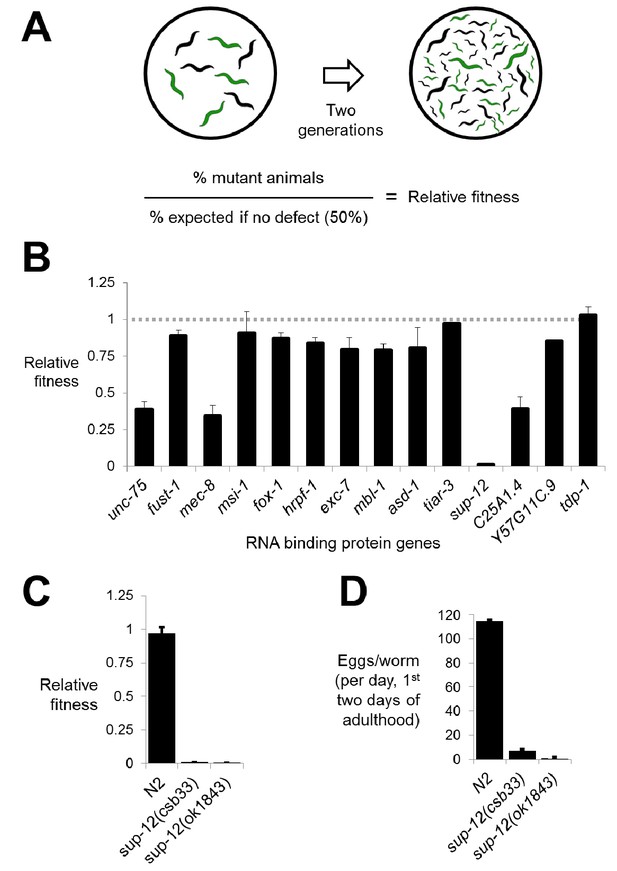
Competitive fitness assays identify RNA binding proteins with important roles in organismal fitness.
(A) Competition assay schematic and calculation of relative fitness as fraction of mutant worms on plate divided by the null expectation of 0.5 assuming no fitness defects in the mutant. (B) Relative fitness values for all single RNA binding protein mutants. (C) Strong fitness deficit in sup-12 mutants confirmed via independent sup-12(ok1843) allele. (D) Egg laying assay reveals fitness defects in sup-12 mutants can be partially explained by greatly reduced fertility. Error bars = S.E.M.
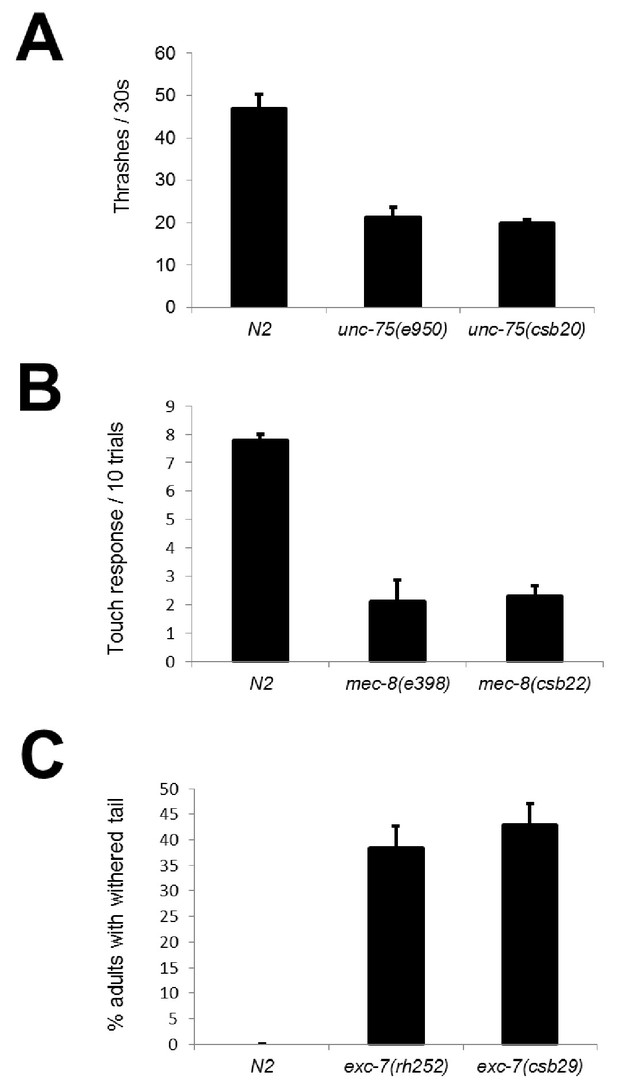
CRISPR mutants recapitulate known RNA binding protein phenotypes.
Comparison between alleles generated in this study and canonical null alleles for (A) uncoordination in unc-75 mutants measured in thrashes per 30 s (B) mechanosensation defects in mec-8 mutants measured as the number of touch responses per 10 trials with an eyelash pick (C) withered tail phenotype in exc-7 mutants show that our CRISPR mutants recapitulate previously-described null phenotypes.Error bars = S.E.M.
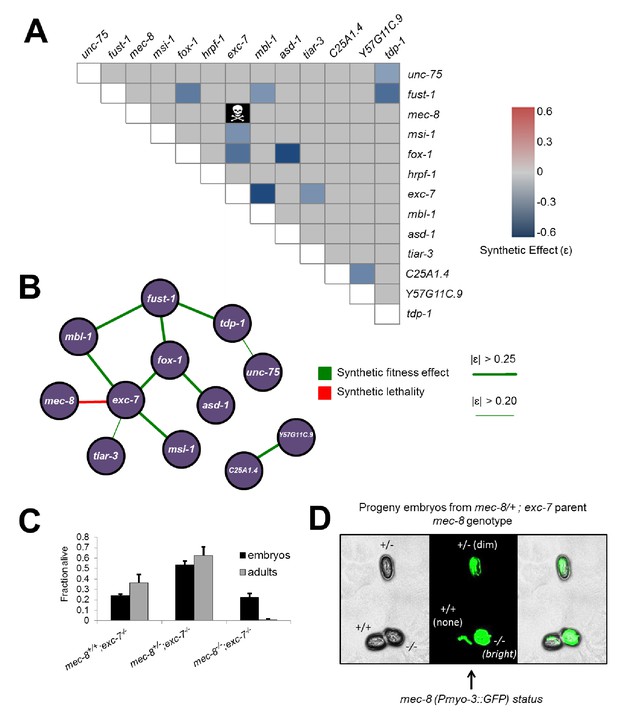
Extensive genetic interactions between RNA binding proteins affecting fitness, viability and lifespan.
(A) Heatmap of synthetic effects for all pairwise genetic interactions (|ɛ| > 0.2). Skull and crossbones denotes synthetic lethality. (B) Network map demonstrating interconnectivity of genetic interactions among RNA binding proteins. Thickness of green edges denotes extent of measured fitness effect. (C) mec-8; exc-7 mutant embyros are laid from mec-8+/−;exc-7 mothers at expected Mendelian ratios, but double homozygotes die during embryogenesis. Error bars = S.E.M. (D) ‘in situ’ genotyping demonstrates that double homozygous embryos are laid at Mendelian ratios. Pharyngeal GFP marks homozygous exc-7 mutation, while body-wall muscle GFP brightness reports on the number of mec-8 mutant alleles (bright = −/−, dim = +/−, none = +/+).
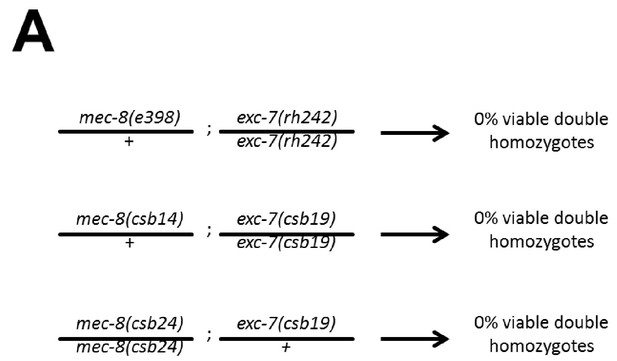
mec-8; exc-7 mutants are synthetic embryonic lethal.
Crosses with CRISPR alleles and canonical alleles confirm that mec-8; exc-7 mutants are synthetically lethal.
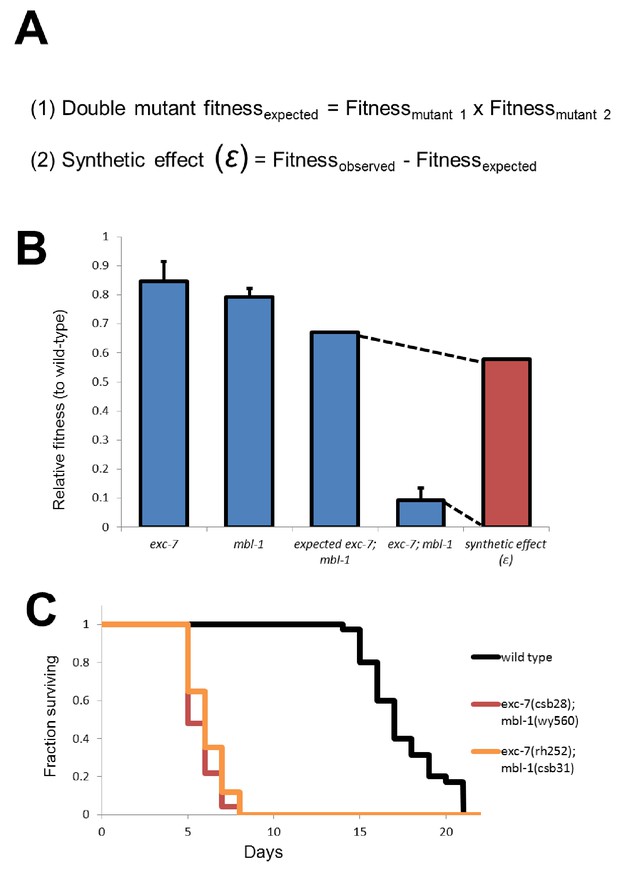
Calculation of synthetic effect (ε) and validation of lifespan defects.
(A) Equations for calculating expected double mutant fitness and synthetic effect. (B) Visual representation using extreme example of exc-7; mbl-1 negative synthetic fitness effect. (C) Lifespan analysis using canonical exc-7 and mbl-1 mutant alleles. Error bars = S.E.M.
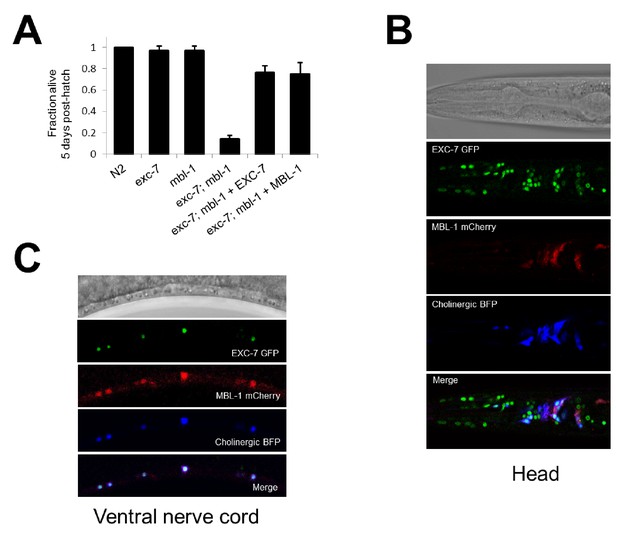
exc-7 and mbl-1 are co-expressed in specific neuronal subtypes.
(A) exc-7; mbl-1 mutants die prematurely and can be rescued by overexpression of either EXC-7 or MBL-1. Error bars = S.E.M.(B) Head region of worm showing exc-7 expression (GFP), mbl-1 expression (RFP) and Cholinergic neurons (BFP). (C) Ventral nerve cord, demonstrating that both RNA binding proteins are co-expressed in the Cholinergic motorneurons.
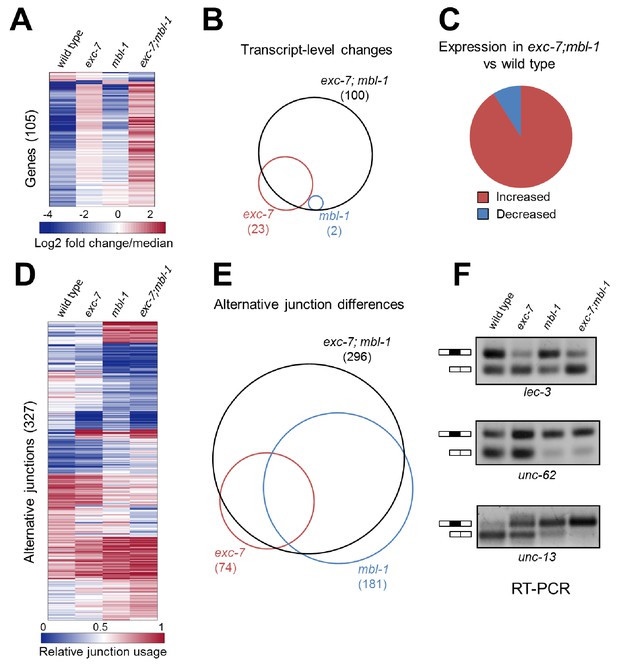
exc-7 and mbl-1 combinatorially control a large network of transcripts and alternative isoforms enriched for genes involved in lifespan regulation.
(A) Heatmap depicting log2 fold change over mean values normalized across each row for wild type and mutant animal samples (columns). (B) Venn diagram depicting number of genes differentially regulated between wild type and mutant strains, and the extent of their overlap. (C) Pie chart depicting bias in expression level changes in exc-7; mbl-1 double mutants relative to wild type animals. (D) Heatmap depicting relative alternative junction usage values for alternatively spliced junctions (rows) across wild type and mutant animal samples (columns). (E) Venn diagram depicting number of differentially regulated alternatively spliced junctions between wild type and mutant strains, and the extent of their overlap. (F) RT-PCR validations for candidate alternative exon skipping events showing changes between wild type and mutant animals. Upper bands represent exon-included isoforms and lower bands represent exon-skipped variants.
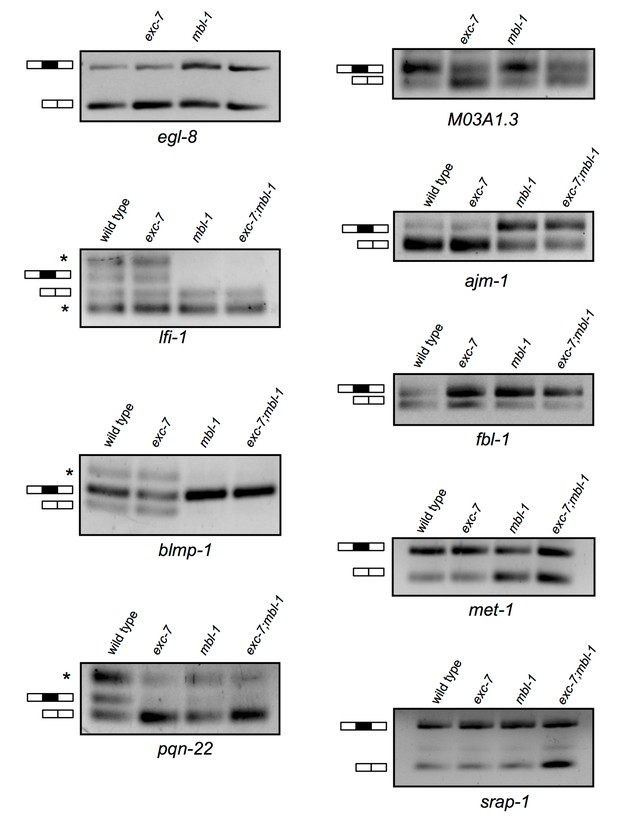
Additional RT-PCRs validating splicing events detected in RNA-Seq analysis.
Whole animal RT-PCRs on RNA extracted from L4 animals. Combined with the RT-PCRs displayed in Figure 4, all (12/12) RT-PCR results agreed with RNA Seq data. Asterisks denote additional larger and/or smaller RNA species that likely correspond to additional unexpected isoforms and/or spurious splicing.
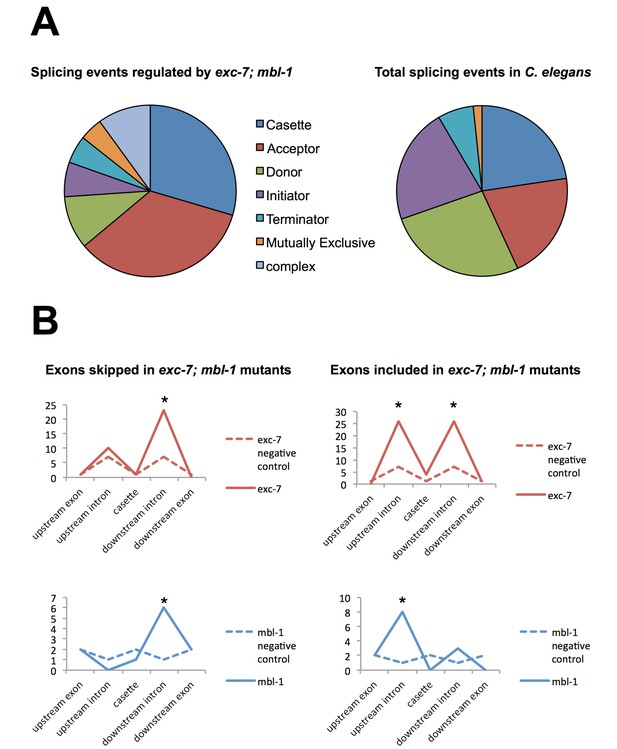
Analysis of alternative splicing events regulated by EXC-7 and MBL-1.
(A) Analysis of alternative splicing events dysregulated in exc-7; mbl-1 mutants shows a large number of affected cassette-type exons and alternative 3’ splice sites (‘splice acceptors’) relative to the reported frequency of these classes of alternative splicing events (frequencies obtained from Ramani et al., 2011). (B) Motif analysis for biochemically-defined cis-elements for EXC-7 or MBNL1 (binding motifs obtained from Ray et al., 2013) near regulated cassette exons. Asterisks denote statistically significant enrichment relative to negative control sequences (Fisher’s exact test, p<0.05).
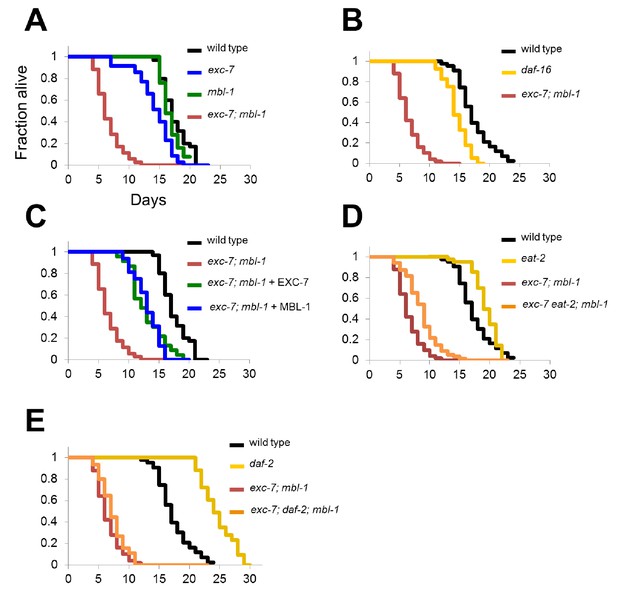
exc-7; mbl-1 mutants have severely shortened lifespans, and genetically interact with the insulin signaling pathway.
(A) exc-7; mbl-1 double mutants, but not single exc-7 or mbl-1 mutants, have strongly reduced lifespans. (B) Lifespan deficits in exc-7; mbl-1 are stronger than the classic short-lived mutant daf-16(mu86). (C) Shortened lifespan in exc-7; mbl-1 mutants can be rescued by transgeneic overexpression of either EXC-7 or MBL-1. (D) eat-2(ad453) mutation increases lifespan in wild-type worms and in exc-7; mbl-1 mutants (p<0.001, log-rank test). (E) daf-2(e1370) mutation strongly increases lifespan in wild-type worms (p<0.001) but not in exc-7; mbl-1 mutants (p>0.05).
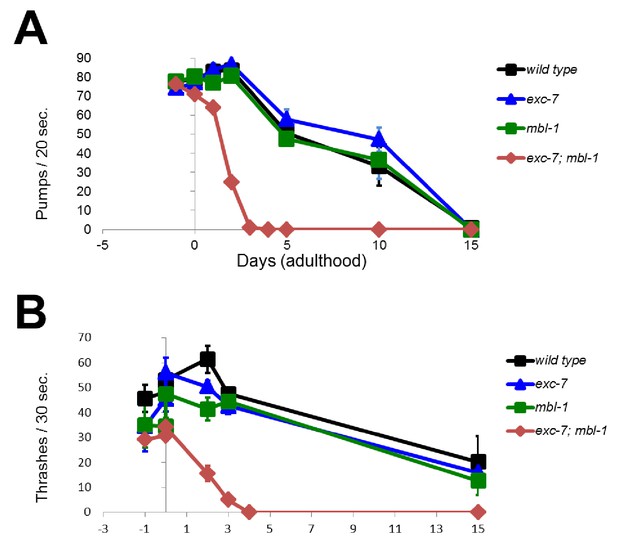
exc-7; mbl-1 mutants appear normal throughout development but have severely shortened adult lifespans.
Behavioral correlates of health such as (A) pharyngeal pumping and (B) locomotion are normal in exc-7; mbl-1 during earlier L4 developmental stage (day −1 of adulthood), but decline quickly upon reaching adulthood. Error bars = S.E.M.
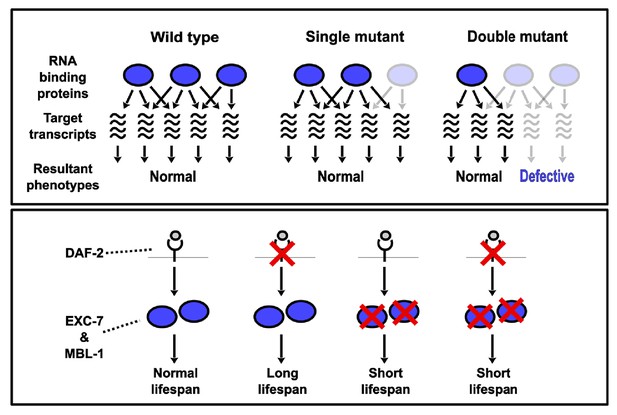
Model depicting synthetic genetic interactions of RNA binding protein genes in this study.
Upper panel displays general model for observed aggravating synthetic genetic interactions between RNA binding protein genes, involving additive or synergistic effects on overlapping sets of target mRNAs. Bottom panel displays model based on our lifespan and epistasis experiments, placing EXC-7 and MBL-1 tentatively downstream of the DAF-2 insulin receptor in regulating lifespan.
Videos
Comparision of wild type and exc-7; mbl-1 double mutant animals.
A movie comparing the gross morphology and movement of wild type animals and exc-7; mbl-1 double mutant animals at 72 hr post-adulthood.
Additional files
-
Supplementary file 1
Genes deleted in this study.
A table displaying the names of the C. elegans genes deleted in this study and their corresponding human orthologs.
- https://doi.org/10.7554/eLife.28129.016
-
Supplementary file 2
Summary of competition experiments.
Table displaying the results of all competition assays performed in this study. First worksheet contains single deletion mutant results, and second worksheet contains double mutant results.
- https://doi.org/10.7554/eLife.28129.017
-
Supplementary file 3
Summary of differentially expressed genes in exc-7 and mbl-1 single mutants, and exc-7; mbl-1 double mutants.
Table displaying the results of our DESeq analysis identifying genes in mutants that are significantly differentially relative to wild type animals.
- https://doi.org/10.7554/eLife.28129.018
-
Supplementary file 4
Summary of differentially utilized alternative junctions.
Table displaying results of our alternative junction analysis comparing wild type, single mutant, and exc-7; mbl-1 double mutant animals. Splice junction chromosome, start and stop coordinates, read counts, junction usage ratios, and p values are listed in the table.
- https://doi.org/10.7554/eLife.28129.019
-
Supplementary file 5
Summary of Gene Ontology (GO) analysis.
Table depicting the lists of statistically enriched GO terms associated with different comparisons to wild type animals (single and double mutant comparisons labeled in spreadsheet). The first worksheet contains analysis on differentially expressed genes, and the second worksheet contains analysis of differentially utilized junctions.
- https://doi.org/10.7554/eLife.28129.020
-
Supplementary file 6
Strains used in this study.
Table displaying information relevant to the mutant animals generated and used in this study. Allele identifiers, genotypes, regions spanning the deletions, and sgRNA targeting sequences used are all listed.
- https://doi.org/10.7554/eLife.28129.021





