Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase
Figures
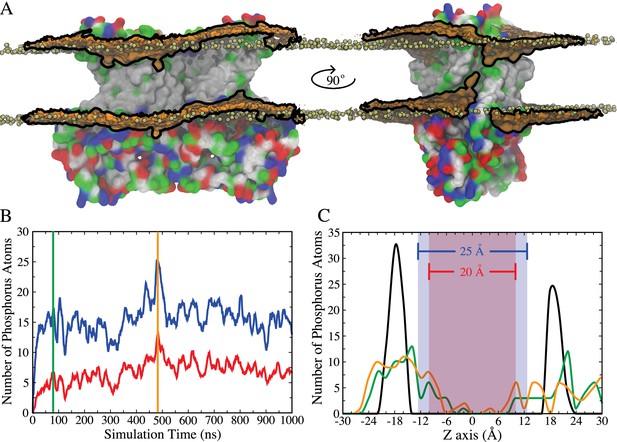
Membrane deformation induced by nhTMEM16.
(A) nhTMEM16 is shown as a molecular surface (extracellular side up) colored by residue type (blue, basic; red, acidic; green, polar; white, nonpolar) in a phospholipid bilayer composed of POPC and POPS. The initial (t = 0) distribution of phospholipids is shown by the tan spheres representing lipid phosphorus atoms. At the end of the 1000-ns MD simulation, the average phospholipid phosphate density within 10 Å of the protein during the Ca2+-activated simulation is shown as an orange surface outlined by black lines that is contoured at 7.5% of the bulk phosphate density. Two different views (rotated by 90° around the membrane normal) are shown, with one of the aqueducts visible in the right panel. (B) Number of phosphorus atoms within 10 Å of the protein that enter the 25 Å- (blue curve) or 20 Å- (red curve) thick core region of the membrane, as defined by the blue and red areas in panel (C). (C) Distribution of phosphorus atoms along the membrane z axis at t = 0 (black curve), t = 80 ns (green curve, corresponding to vertical green bar in panel (B)), and t = 480 ns (orange curve, corresponding to vertical orange bar in panel (B)).

nhTMEM16 surface hydrophobicity.
Molecular surface representation of nhTMEM16 crystal structure (4WIT), colored according to the hydrophobicity scale of Kyte and Doolittle (Cyan = hydrophilic (−4.5), Orange = hydrophobic (4.5)), illustrating the hydrophilic surfaces near the entrances of the aqueduct (blue arrows), exposed to the hydrophobic core of the membrane. The aqueduct is lined by transmembrane helices TM4 and TM6. The membrane bilayer is represented by dashed lines.
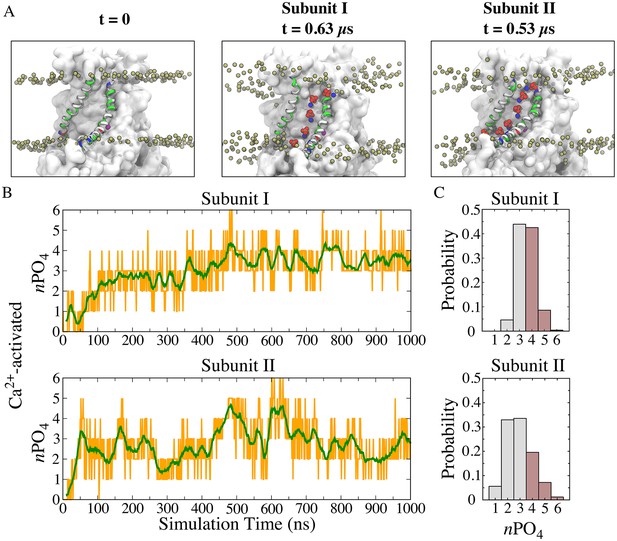
Features of the ‘aqueduct’ on nhTMEM16 surface.
(A) Representative snapshots of the aqueduct during an MD simulation with Ca2+-activated nhTMEM16. Aqueduct-lining helices TM4 and TM6 are colored by residue type. Phospholipid head groups in the bilayer are shown by tan phosphorus atoms while those inside the aqueduct are shown by phosphorus (tan), oxygen (red) and nitrogen (blue) van der Waals atoms. Ca2+ ions are purple spheres. The aqueduct is devoid of phospholipid head groups at t = 0 (left panel), but becomes occupied by lipids in both subunits during the simulation. (B) Number of phosphate groups (orange trace) and the moving average (green, bin = 20) within the 20 Å-thick core region. (C) Normalized probability histograms of the phosphate count in the 20 Å-thick core region of each aqueduct during the last 500 ns. Numbers larger than three are highlighted in brown.
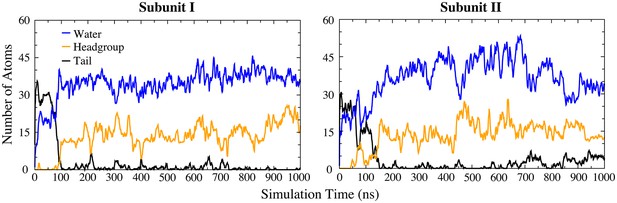
Penetration of lipids into the aqueduct in the Ca2+-activated simulation.
Number of water oxygen atoms (blue), lipid head group heavy atoms (orange), and lipid tail carbon atoms (black) within the aqueduct are plotted vs. simulation time. Within the first 100–150 ns, lipids penetrate the aqueduct (decrease in tails (black) and increase in head groups (orange)). This is accompanied by an increased hydration (water, blue).
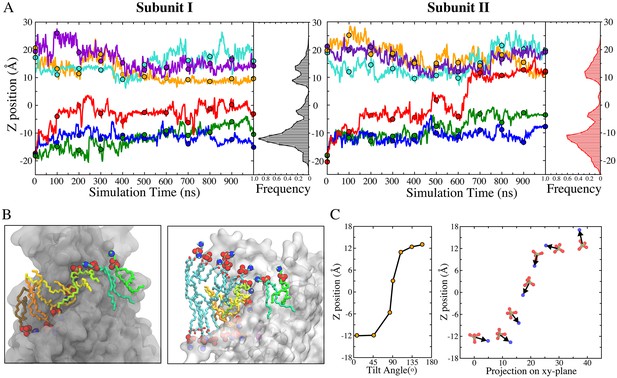
Kinetics of lipid permeation.
(A) Translocation of lipids along the aqueduct measured as the z-positions of phosphorus atoms versus time. Each 100-ns time point is highlighted as a dot. Complete flipping of a lipid from the inner leaflet to the outer leaflet is captured in subunit II (red trace). Histograms plot the frequency of phosphorus atoms in 1-Å bins along the aqueduct. (B) Views showing representative lipids in the aqueduct. Left panel: Front view with hydrocarbon chains of each phospholipid inside the aqueduct colored differently. Right panel: same image rotated 90o with added phospholipids in proximity to the aqueduct colored with cyan tails. The head groups of these lipids are organized in a curve with the lipids closest to the aqueduct being located closer to z = 0. (C) Left: representative orientations of lipid tails along the aqueduct. The tilt angle of the lipid tail is defined by the angle bisector vector of the two acyl tails and z axis. Right: corresponding P-N dipole orientations of the head groups along the aqueduct.
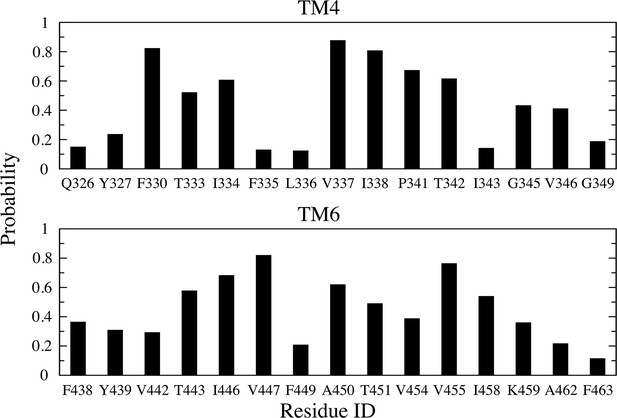
Fraction of time each amino acid forms hydrophobic contact (<4 Å) with a fatty acid acyl chain whose head group is inside the aqueduct.
Analysis is combined for the two subunits. Residues with >10% time in contact are shown.
Full scrambling of a POPC lipid from inner leaflet to outer leaflet of the membrane through the aqueduct under equilibrium condition.
One subunit of the nhTMEM16 dimer is shown colored N- to C- terminus blue to red. Ca2+ ions are purple spheres. The simulation is 950 ns in duration. View from the plane of the membrane. The aqueduct is to the left and the dimer interface to the right.
Full scrambling of a POPC lipid from inner leaflet to outer leaflet of the membrane through the aqueduct under equilibrium condition.
One subunit of the nhTMEM16 dimer is shown colored N- to C- terminus blue to red. Ca2+ ions are purple spheres. The simulation is 950 ns in duration. View looking into the aqueduct from the plane of the membrane.
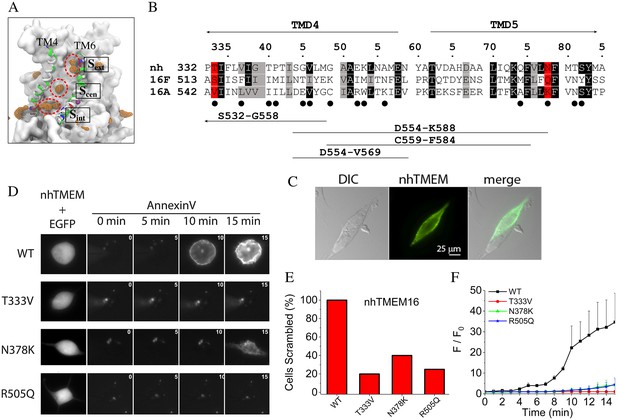
Lipid binding sites and effects of mutations in nhTMEM16.
(A) Volumetric map of phosphate occupancy extracted from the Ca2+-activated simulation is shown as orange wireframe contoured at isovalue 0.15 overlaid on the protein structure. The three phosphate binding sites (circled and labeled) are characterized in detail in Figure 4—Figure supplement 1. (B) Alignments of TM4 and TM5 of nhTMEM16 (nh), mouse TMEM16F (16F), and mouse TMEM16A (16A). Structure-based alignments were performed using PROMALS3D. The sequences of nhTMEM16 and the mammalian proteins are 31% similar over the aligned region. Backgrounds are colored to indicate identity (black) or similarity (grey). nhTMEM16 amino acids that directly interact with lipid head groups for >10% of the time are indicated with a filled circle (●). Amino acids that interact with lipid head groups and are conserved between nhTMEM16 and TMEM16F but different in TMEM16A are highlighted in red. Consensus amino acid numbering in the top line refers to nhTMEM with the first residue numbers preceding the sequence. At the bottom, the four horizontal lines indicate TMEM16A sequences that were previously replaced with TMEM16F sequences to convert TMEM16A into a phospholipid scramblase (Yu et al., 2015). (C) 3X-FLAG-nhTMEM16 is trafficked to the plasma membrane of HEK-293 cells. Cells were transiently transfected with a synthetic codon-optimized nhTMEM16 tagged on the N-terminus with 3X-FLAG. Cells were fixed and stained with anti-FLAG antibody followed by a fluorescent secondary antibody and imaged with a wide-field microscope. Left: differential interference contrast image. Middle: immunofluorescence. Right: merged image. Two attached cells are shown. One cell was transfected with nhTMEM16 and one was not transfected. (D) Left: effects of mutations in nhTMEM16 on phospholipid scrambling. Cells were co-transfected with plasmids encoding nhTMEM16 (WT or mutant) and EGFP. Transfected cells were identified by green fluorescence (left panel). Phospholipid scrambling was measured by binding of Annexin-V-AlexaFluor-568 at 0, 5, 10, and 15 min after establishing whole-cell recording with an internal solution containing 200 µM free Ca2+ (right panels). (E) Percentage of cells binding Annexin-V above background within 15 min after establishing whole cell recording with a high Ca2+ internal solution. Each bar represents 4 to 5 cells. (F) Time course of phospholipid scrambling by WT and mutant nhTMEM16.

Amino acid residue interaction with lipid head groups in the aqueduct.
(A) Coordinating residues surrounding each binding site are labeled. The protein ribbon is colored based on the frequency of each residue forming electrostatic interaction with the head groups inside the aqueduct during the simulation. The darkest blue color represents residues interacting with lipids > 30% of the time. Analysis is combined for the two subunits. (B) Data showing the residues with >10% time of interaction. Residues that are targeted for mutations are shown in red.
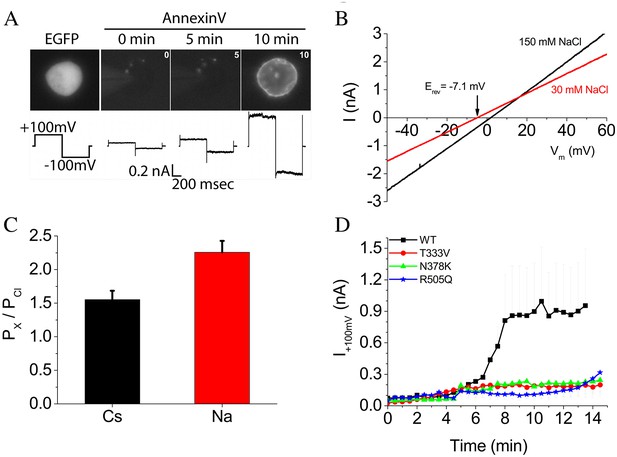
Phospholipid scrambling and ionic currents in nhTMEM16.
(A) Phospholipid scrambling and ionic currents are stimulated in HEK cells transfected with nhTMEM16 and co-transfected with a plasmid encoding EGFP. Transfected cells were identified by green fluorescence. Phospholipid scrambling was measured by binding of Annexin-V-AlexaFluor-568 at 0, 5, and 10 min after establishing whole-cell recording with an internal solution containing 200 µM free Ca2+. Ionic currents recorded by a voltage step from a holding potential of 0 mV to +100 mV followed by a step to −100 mV are shown below each Annexin-V fluorescent image. Untransfected HEK cells do not scramble in the 15 min time frame as previously shown (Yu et al., 2015). (B) Ionic selectivity of WT nhTMEM16 currents. Current-voltage relationships were measured using voltage ramps 1 s duration from −100 mV to +100 mV. Ionic selectivity was determined by the change in reversal potential measured in response to switching between extracellular solutions containing 150 mM NaCl (black curves) and 30 mM NaCl (red curves). (C) Relative permeability of Na+ and Cs+ to Cl- (PNa:PCl and PCs:PCl) calculated from the Goldman-Hodgkin-Katz equation. (D) Time course of development of ionic currents at +100 mV by WT and mutant TMEM16.
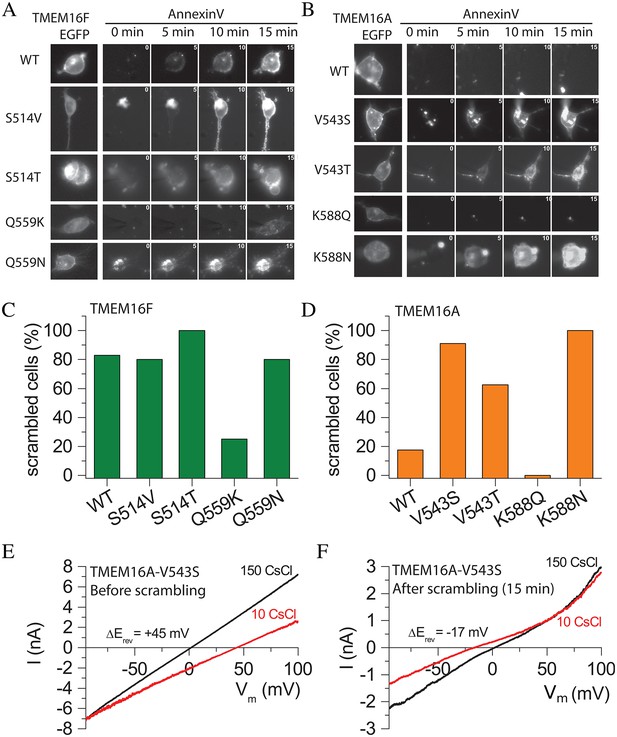
Effects of amino acid mutations on TMEM16F (A,C) and TMEM16A (B,D).
(A and B) Images of cells during phospholipid scrambling. Single cells were patch clamped and intracellular Ca2+ controlled by 200 µM Ca2+ in the pipet. The first image in each row is EGFP fluorescence of the tagged TMEM16 protein. The next images are Annexin-V fluorescence taken 0, 5, 10, and 15 min after raising Ca2+ by initiating whole cell recording. (C and D) Percentage of cells binding Annexin-V within 15 min after establishing whole cell recording with a high Ca2+ internal solution. Each bar represents between 5 and 29 cells. (E and F) The V543S mutation in TMEM16A alters ionic selectivity. Current-voltage relationships were measured using voltage ramps of 1 s duration from −100 mV to +100 mV. Ionic selectivity was determined by the difference in reversal potential between extracellular 150 mM CsCl (black curves) and 10 mM CsCl (red curves). (E) Switching between 150 mM and 10 mM CsCl immediately after establishing whole-cell recording shifted Erev +45 mV, corresponding to a PCl:PCs = 9.7, whereas (F) switching after Annexin-V binding had reached a plateau, Erev shifted −17 mV in the opposite direction, corresponding to PCl:PCs = 0.45.
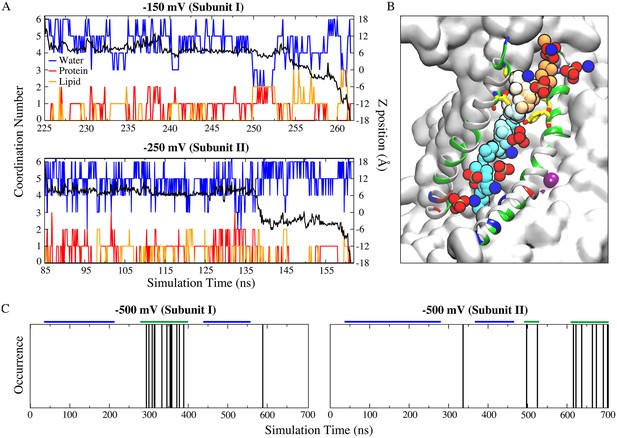
Ion permeation occurs through the same structural pathway of lipid scrambling.
(A) Full permeation of one Na+ ion from the extracellular side to the intracellular side of the membrane at −150 mV (top panel) and −250 mV (bottom panel). Coordination number (left y-axis) and z-position (right y-axis) of the permeant Na+ ions as a function of time. Oxygen atoms from protein (red) and lipid head groups (orange) both contribute to the ion coordination throughout their permeation (−150 mV: ~36.7 ns duration starting at ~225 ns; −250 mV: ~76.4 ns duration starting at ~85 ns). The coordination number for water oxygen atoms is blue. The black curve is the corresponding z-position of the permeant ion. (B) Time series snapshots representing translocation of the Na+ ion with time represented in pseudo-color from orange (start) to cyan (end). Y439 and T333 are shown in stick representations. The lipid head groups lining the ion permeation pathway are shown as phosphate groups and nitrogen atoms. (C) Multiple ion permeation events occur in both subunits at −500 mV during the 700 ns simulation. Each ion permeation event is indicated by a vertical line at the time point when the permeation is complete. The segments marked by the blue and green bars were used for measuring the center of mass distances between the aqueduct lining helices TM4 and TM6 as shown in Figure 7—figure supplement 4.

Translocation of the permeant Na+.
(A and B) Left panels: Translocation of the permeant Na+ (at −150 mV (A) or −250 mV (B)) through the aqueduct, measured as its z-position versus permeation time. The coordinating residues during ion permeation are shown as dots color-coded to the amino acid residues in the inset. Right panels: The proportion of coordination shows the contribution of each participating residue during the ion permeation.

Na+ permeation at −500 mV.
(A) Distribution of Na+ permeation duration for the 24 permeation events at −500 mV. One third of the events took place within five ns. (B). The proportion of coordination shows the contribution of each participating residue during the ion permeation events.
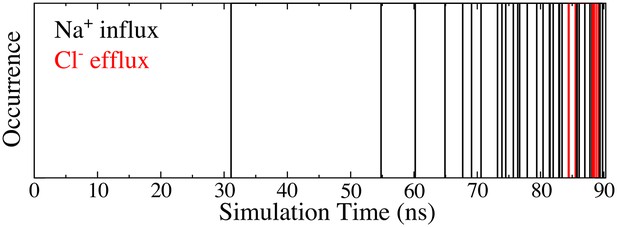
At higher voltages (−1 V), both Na+ influx (38 occurrences) and Cl- efflux (eight occurrences) were observed during a 90 ns simulation.
https://doi.org/10.7554/eLife.28671.017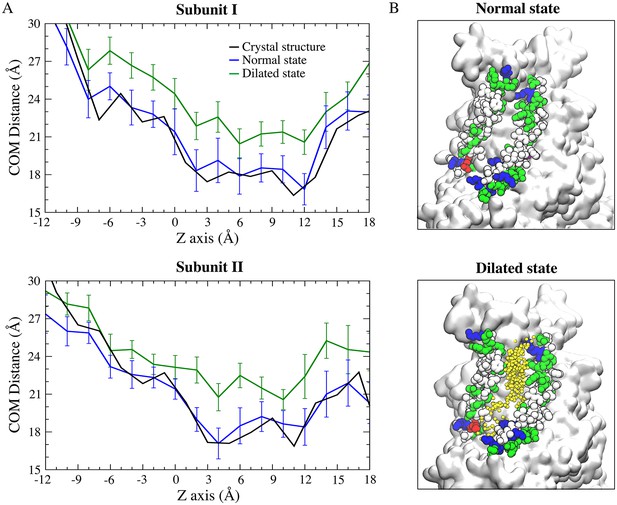
Dilation of the aqueduct.
(A) Average center of mass (COM) distance between TM4 and TM6 in the normal (blue) and dilated (green) states for each subunit during the 700 ns simulation at −500 mV. The COM distance for the normal state is similar to that of the crystal structure (black). (B) Representative snapshots of the normal (top panel) and dilated (bottom panel) states of the aqueduct for ion conduction. Residues in TM4 and TM6 are shown as van der Waals and colored by residue type (blue, basic; red, acidic; green, polar; white, nonpolar). The remaining part of the protein is shown as white molecular surface. Representative footprints of the permeant Na+ ions are shown as yellow dots in the dilated state.
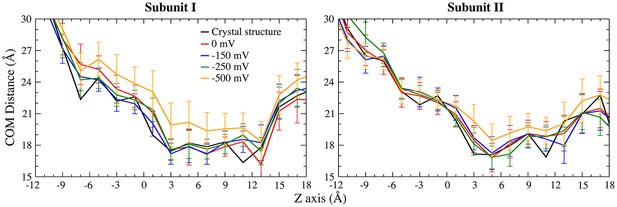
Comparison of the aqueduct conformation with different transmembrane potentials.
The average COM distances between TM4 and TM6 at −150 mV and −250 mV are similar to that of the crystal structure and simulation at 0 mV; the value at −500 mV is slightly larger, due to the transient dilation phase.
Movie shows part of the simulation trajectory (~200 to 400 ns) at −500 mV, during which multiple ion permeations were captured through the aqueduct in subunit I.
Phospholipid phosphorus atoms are tan, Na+ ions are red. TM4 and TM6 are colored by residue type.
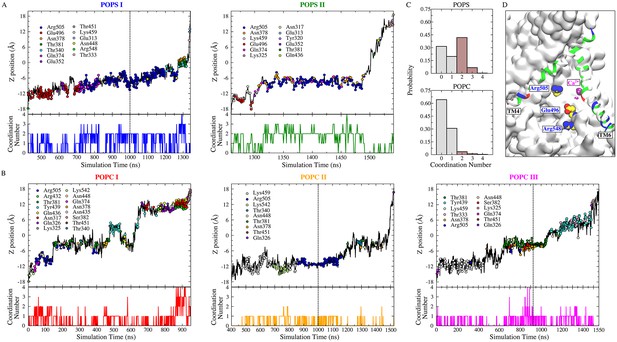
POPS is more coordinated by protein than POPC during flopping.
(A) Full permeation of POPS from the inner leaflet to the outer leaflet was captured in subunit I (left) and II (right) during the 1700 ns simulation (1000 ns equilibrium followed by 700 ns simulation with −500 mV transmembrane potential). The dashed line indicates the time of electric field application. Top panels: Translocation of POPS through the aqueduct, measured as the z-position of its phosphorus atom versus time. The head group coordinating residues during lipid flopping are shown as dots color-coded to the amino acid residues in the inset. Bottom panels: Head group coordination number of the scrambled POPS as a function of time. (B) Full permeation of POPC. POPC I (red) was fully scrambled before the external electric field was applied. (C) Normalized probability histograms of the head group coordination number during the lipid scrambling, averaged for the scrambled POPS (top panel) and POPC (bottom panel). POPC is less coordinated by the protein compared to POPS. (D) Residues inside the aqueduct that strongly favor the binding of POPS over POPC are labeled (E496, R505, and R548).
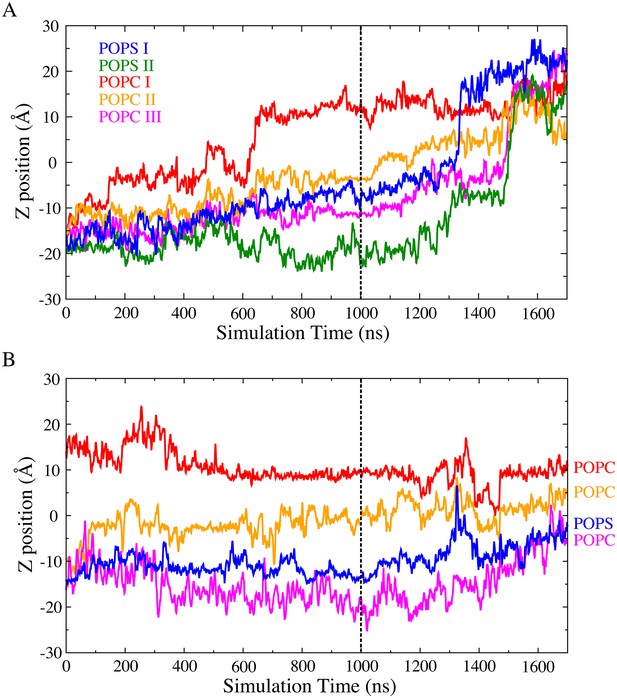
Scrambling events captured during the 1700 ns simulation.
(A) Multiple full scrambling for POPS and POPC from the inner leaflet to the outer leaflet were captured in both subunits. Translocation of the scrambled lipid through the aqueduct was measured as the z-position of its phosphorus atom versus time. The dashed line indicates the time point of adding external electric field. (B) Half-scrambling events captured in subunit I. 2 POPC (orange and magenta curves) and 1 POPS (blue curve) were observed to pass the membrane midplane from the inner leaflet. 1 POPC (red curve) was observed to pass the membrane midplane from the outer leaflet.
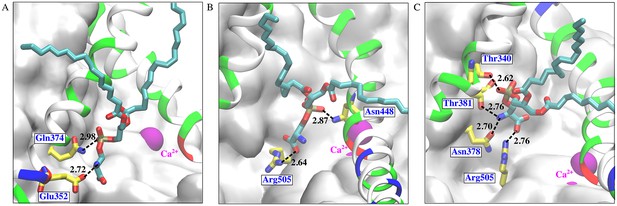
POPS binding inside the aqueduct.
Representative snapshots showing the phosphate and serine of the POPS head group are both coordinated by residues inside the aqueduct during the scrambling. POPS is coordinated by interactions between (A) Q374, E352 and head group PO4 and NH3, (B) R505 and N448 and head group PO4 and COO-, and (C) T340, T381, N378, R505 and head group PO4, NH3 and COO-. Dashed line represents the coordination distance (Å). Oxygen atoms are red, nitrogen blue and phosphorus tan. Carbon atoms in amino acids are yellow and are cyan in lipid. TM4 and TM6 are shown as helices colored by residue type. The remaining part of the protein is shown as white molecular surface. Ca2+ ions are purple spheres.
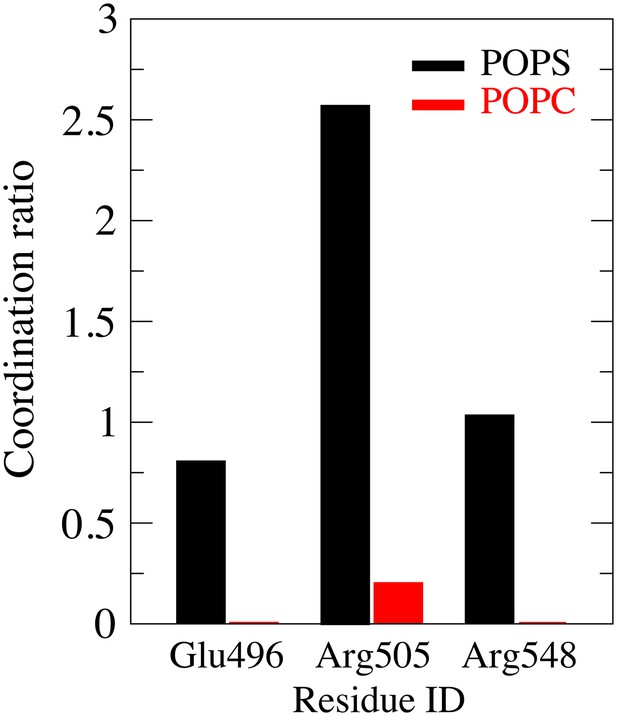
Comparison of POPS and POPC head group coordination from residues E496, R505 and R548 during the 1700 ns simulation.
https://doi.org/10.7554/eLife.28671.024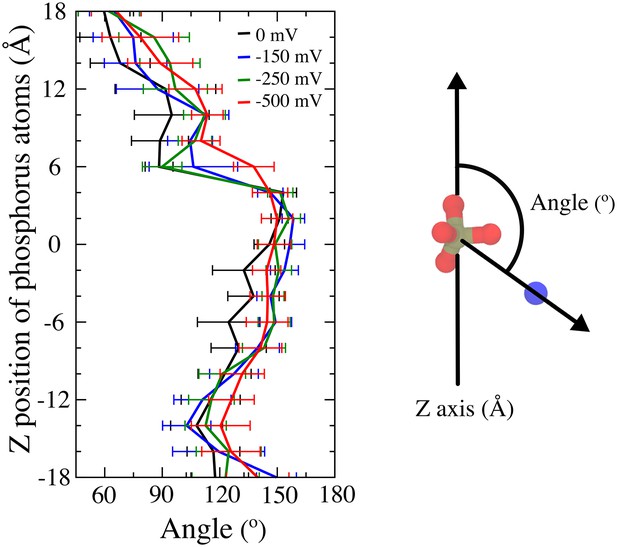
Lipid P-N dipole orientations for head groups inside the aqueduct (bin = 2 Å along z axis) over the simulations with different transmembrane potentials.
The angle is defined by the P-N vector with respect to z axis. The phosphate groups precede the choline (POPC) or amino (POPS) groups in the lower half of the aqueduct (up to 4 Å) with obtuse angles respect to z axis. Transition of obtuse angles to right angle was observed at ~5 Å. When the phosphate groups move above 12 Å along the aqueduct, the angles become acute and the phosphate groups thus succeed the choline or amino groups.
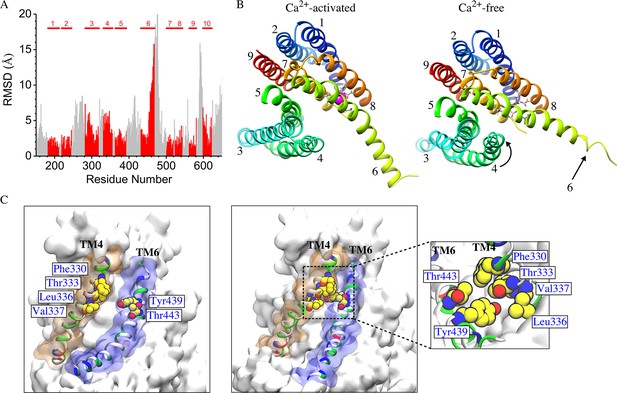
Difference between Ca2+-free and Ca2+-liganded nhTMEM16.
(A) Ca2+-free and Ca2+-liganded nhTMEM16 were structurally aligned with UCSF Chimera and the RMSD of each residue calculated using least squares. TM domains are indicated at the top and RMSD values for TM domains are colored red. (B) Representative structures for Ca2+-activated (at ~837 ns) and Ca2+-free (at ~487 ns) nhTMEM16 from the 1 µsec simulations, respectively. The TM domains are colored and labeled. Ca2+ ions are magenta. Ca2+-coordinating residues are represented as sticks. (C) Illustration of the aqueducts in (B) showing the normal conformation (left panel) and the pinching near the gating region in the Ca2+-free state (right panel). Pinching occurs when TM4 and TM6 are in close contact (sidechain-sidechain distances < 3.0 Å) near the bottleneck region. TM4 and TM6 are colored by residue type and contoured by orange and blue molecular surfaces. Residues involved in inter-helical contacts (F330, T333, L336 and V337 from TM4, Y439 and T443 from TM6) are shown as van der Waals spheres. The inset shows the top view of the pinching of the aqueduct by the hydrophobic contacts of residues from TM4 and TM6.
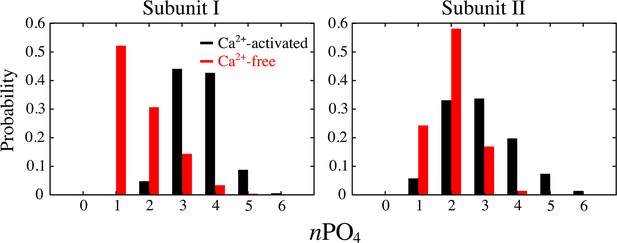
Comparison of head group distributions in the Ca2+-activated and Ca2+-free simulations.
Normalized probability histograms of the phosphate count in the 20 Å-thick core region of the aqueduct in subunit I (left panel) and II (right panel) were calculated for the last 500 ns of the simulations.
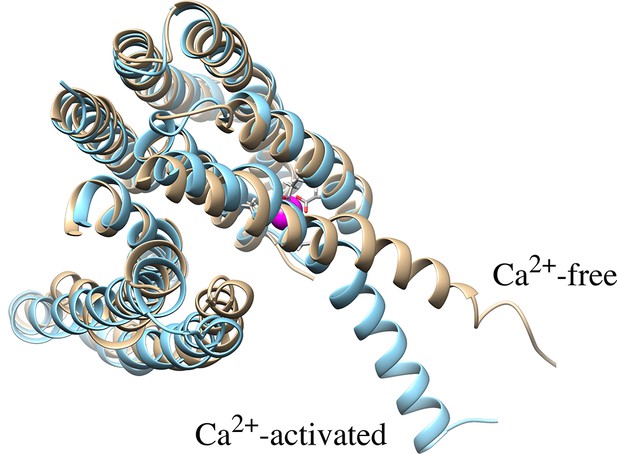
Overlaid structures of Ca2+-free (tan) and Ca2+-activated (sky blue) nhTMEM16 as shown inFigure 9.
Ca2+ ions are magenta. Ca2+-coordinating residues are represented as sticks.
Movie shows a morph between the representative conformations of the Ca2+-free and Ca2+-activated states.
Gating residues F330, T333, L336, V337, Y439, and T443 are shown in spacefilled with atoms colored by element (red, oxygen; grey, carbon; white, hydrogen). The morph was created in UCSF Chimera 1.11.2.
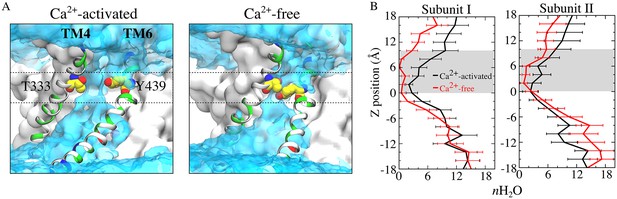
Aqueduct hydration.
(A) Average water density of the Ca2+-activated (left panel) and Ca2+-free (right panel) simulations is shown as a transparent cyan surface contoured at 0.15 of bulk water density, overlaid on representative snapshots of each aqueduct. Gating residues T333 and Y439 are shown as van der Waals spheres. The gating region of the aqueduct (0 < z < 10 Å, membrane center z = 0) is highlighted by dashed lines. (B) Average number of water molecules during the last 150 ns of the 1000 ns of the Ca2+-activated and Ca2+-free simulations. The grey area represents the gating region.
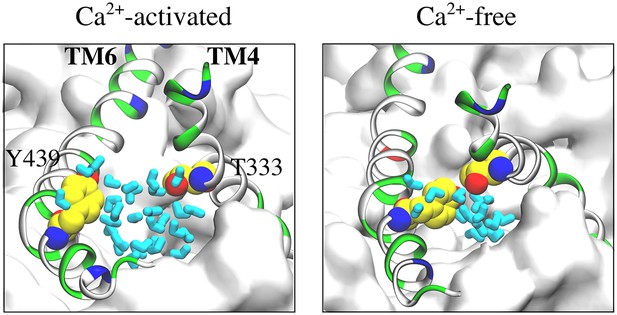
Gating region of the aqueduct.
Representative snapshots showing the top view of the gating region of the aqueduct (z > 10Å is removed for clarity) during the Ca2+-activated (left panel) and Ca2+-free (right panel) simulations. Water molecules in this region are colored in cyan.
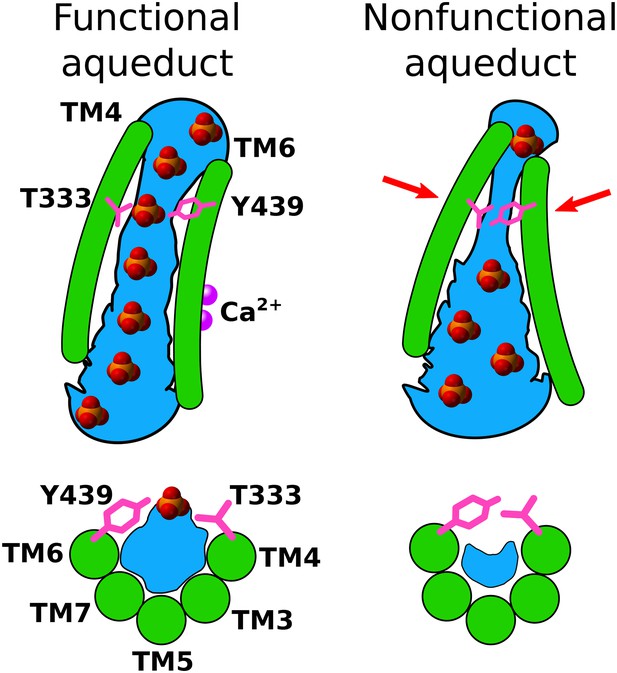
A model for Ca2+-activated lipid scrambling.
Top panels: view from the membrane. Bottom panels: view from extracellular space. Left: Ca2+-activated. Right: Ca2+-free. Hydration is shown in blue and Ca2+ ions are shown as purple spheres. In the presence of Ca2+, a hydrated pathway for lipid head groups is open, but in the absence of Ca2+, tight contact between gating residues T333 and Y439 narrows the aqueduct, resulting in poor hydration in the gating region and prevents head groups from passing through the aqueduct.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28671.033





