Identification of highly-protective combinations of Plasmodium vivax recombinant proteins for vaccine development
Figures
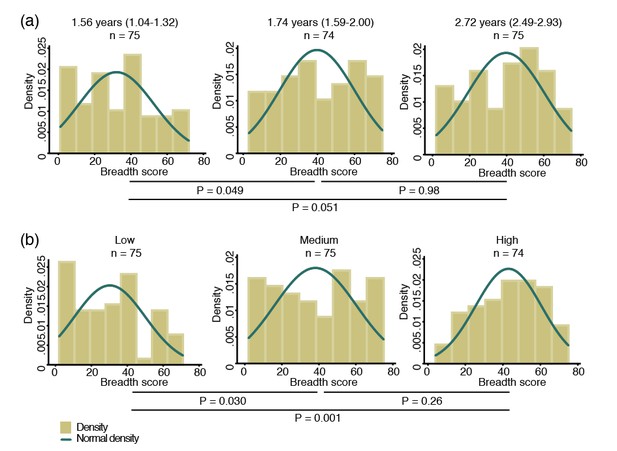
Breadth of IgG antibodies to 38 P. vivax proteins in Papua New Guinean children aged 1–3 years.
For each protein, antibody levels were stratified into tertiles and scored as 0, 1 or 2 for the low, medium, and high tertiles, respectively. Scores were then added up to reflect the breadth of antibodies per child. (a) Antibody breadth by age group. Age is shown as median (interquartile range). (b) Antibody breadth by lifetime exposure group. For each child, exposure was defined as the total number of P. vivax blood-stage clones acquired per time-at-risk (molFOB), and lifetime exposure as a product of age and molFOB. P values are from negative binomial regression and were deemed significant if <0.05.
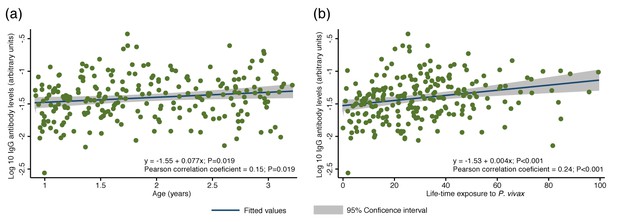
Association between cumulative IgG levels to 38 P. vivax proteins and exposure to P. vivax infections in Papua New Guinean children aged 1–3 years.
(a) Association with age. (b) Association with lifetime exposure. For each child, exposure was defined as the total number of P. vivax blood-stage clones acquired per time-at-risk (molFOB), and lifetime exposure as a product of age and molFOB. n = 225. P values < 0.05 were deemed significant.
-
Figure 2—source data 1
Associations between IgG to P. vivax antigens with measures of concurrent and cumulative exposure in Papua New Guinean children aged 1–3 years.
Geom mean = geometric mean. Exposure was defined as the total number of P. vivax blood-stage clones acquired per time-at-risk (molFOB) and lifetime exposure as age multiplied by molFOB. Geometric mean IgG levels and 95% intervals are in arbitrary units interpolated form standard curves using a 5PL logistic regression model and were multiplied by 1000. For age and lifetime exposure, rho (r) and P values were calculated using Spearman's rank test. For infection status, IgG levels were log10 transformed and P values calculated using unpaired 2-tailed t tests. P values < 0.05 were deemed significant.
- https://doi.org/10.7554/eLife.28673.005
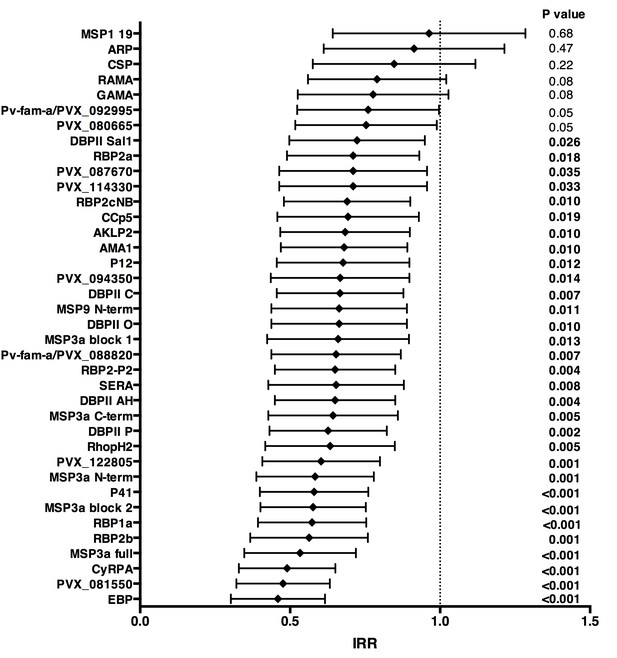
Association between high IgG levels to 38 P. vivax proteins and protection against clinical malaria (density >500/μL) in Papua New Guinean children aged 1–3 years old.
Data are plotted as incidence rate ratios and 95% confidence intervals adjusted for exposure (molFOB), age, season, and village of residency. Incidence rate ratios are for high versus low tertiles of responders, 95% confidence intervals and P values are from GEE models. P values < 0.05 were deemed significant.
-
Figure 3—source data 1
Associations between antibodies to 38 P. vivax proteins and risk of P. vivax clinical episodes (>500 parasites/μL) in Papua New Guinean children aged 1–3 years.
M = medium IgG levels; H = high IgG levels; uIRR = Unadjusted incidence rate ratio; aIRR = Adjusted incidence rate ratio, adjusted for exposure (molFOB), age, village of residency, and season. P values were from GEE models and were deemed significant if <0.05.
- https://doi.org/10.7554/eLife.28673.007
-
Figure 3—source data 2
Associations between antibodies to 38 P. vivax proteins and risk of P. falciparum clinical episodes (>2500 parasites/μL) in Papua New Guinean children aged 1–3 years.
M = medium IgG levels; H = high IgG levels; uIRR = Unadjusted incidence rate ratio; aIRR = Adjusted incidence rate ratio; adjusted for exposure (molFOB), age, village of residency and season. P values were from GEE models and were deemed significant if <0.05.
- https://doi.org/10.7554/eLife.28673.008
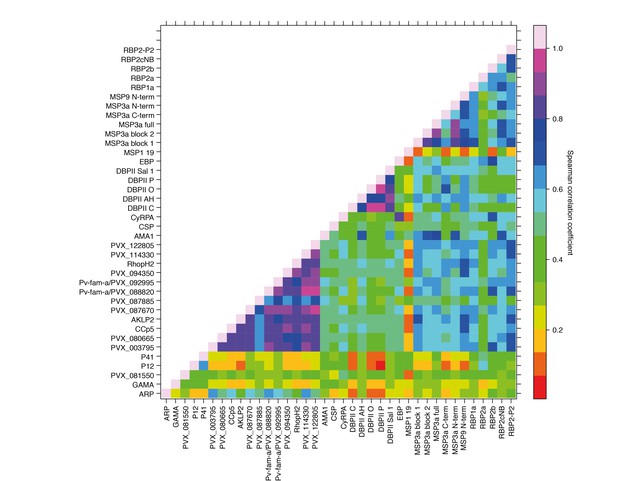
Correlations between IgG to 38 P. vivax proteins in Papua New Guinean children aged 1–3 years.
Correlation coefficients between antibody levels to every pair of antigens were calculated using Spearman’s rank correlation tests.
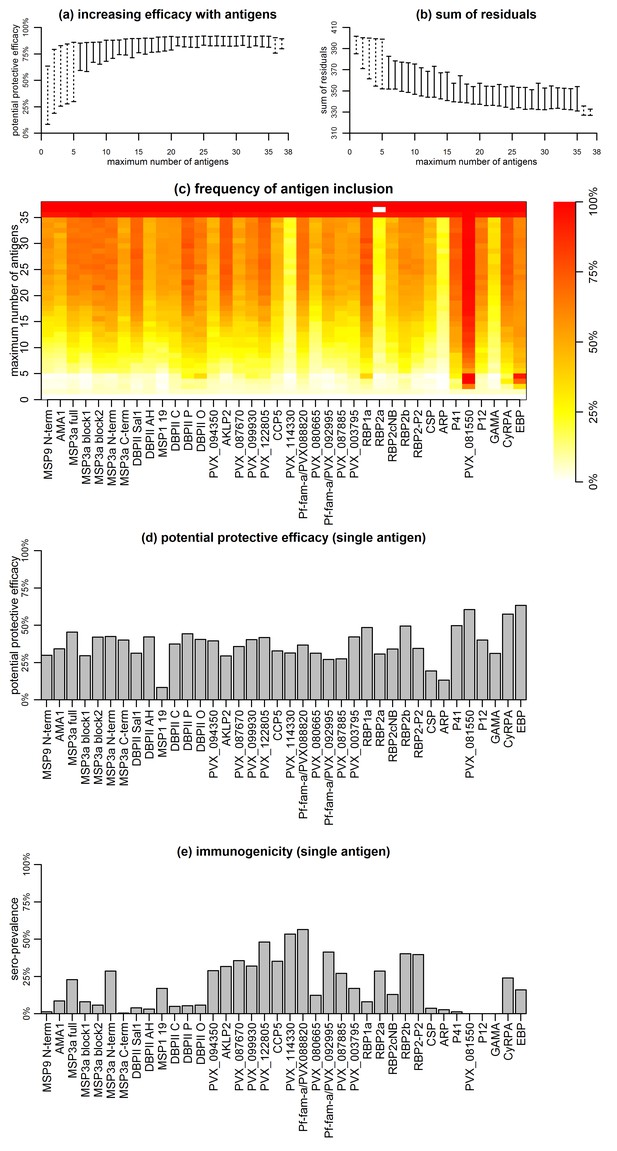
Association between antibodies to combinations of P. vivax proteins and malaria risk in Papua New Guinean children aged 1–3 years.
(a) Potential protective efficacy (PPE) for combinations of antigens with the maximum number of antigens indicated on the x-axis. Dashed lines represent the range of PPE for all possible combinations. Solid lines represent the range of PPE from 1000 implementations of the simulated annealing algorithm. (b) Sum of residuals (as a measure of model goodness of fit). (c) The heatmap shows the frequency of including an antigen (x-axis) in a multi-component vaccine with a fixed number of antigens (y-axis). (d) Predicted PPE of a single antigen. (e) Immunogenicity of each antigen represented as seroprevalence with a cut-off as 10% of the antibody levels of fully-immune PNG adults.
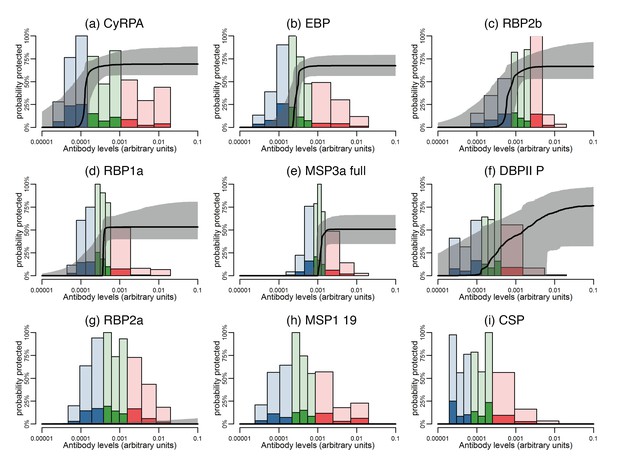
Estimated dose-response curves for the associations between antibody responses specific to P. vivax antigens and protection from clinical malaria.
Solid black lines depict exposure-adjusted dose-response curves, and the grey shaded regions depict the 95% credible intervals. Histograms show the observed distribution of antibody levels (relative to the PNG immune pool) colored per tertiles (low = blue; medium = green; high = red), and the darkly-colored portions of the histograms show the proportion of individuals with that antibody level who had a P. vivax episode (>500 parasites/μL). (a–c) Examples of antigens that need low antibody levels to provide 50% of protection. (d–f) Examples of antigens that need medium antibody levels to provide 50% of protection. (g–i) Examples of antigens that need high antibody levels to provide 50% of protection.
-
Figure 6—source data 1
Antibody levels and 50% protection from clinical malaria.
AB50 is the estimated antibody levels associated with a 50% probability of protection from clinical malaria. Antibody levels are measured relative to a 1:100 dilution from a pool of plasma from immune Papua New Guinean adults. Estimates of AB50 are also presented as a proportion of the immune plasma pool. The maximum protection is the protection at an antibody level of 0.01. The median of the correlations of that antigen with the other 37 antigens is presented.
- https://doi.org/10.7554/eLife.28673.013
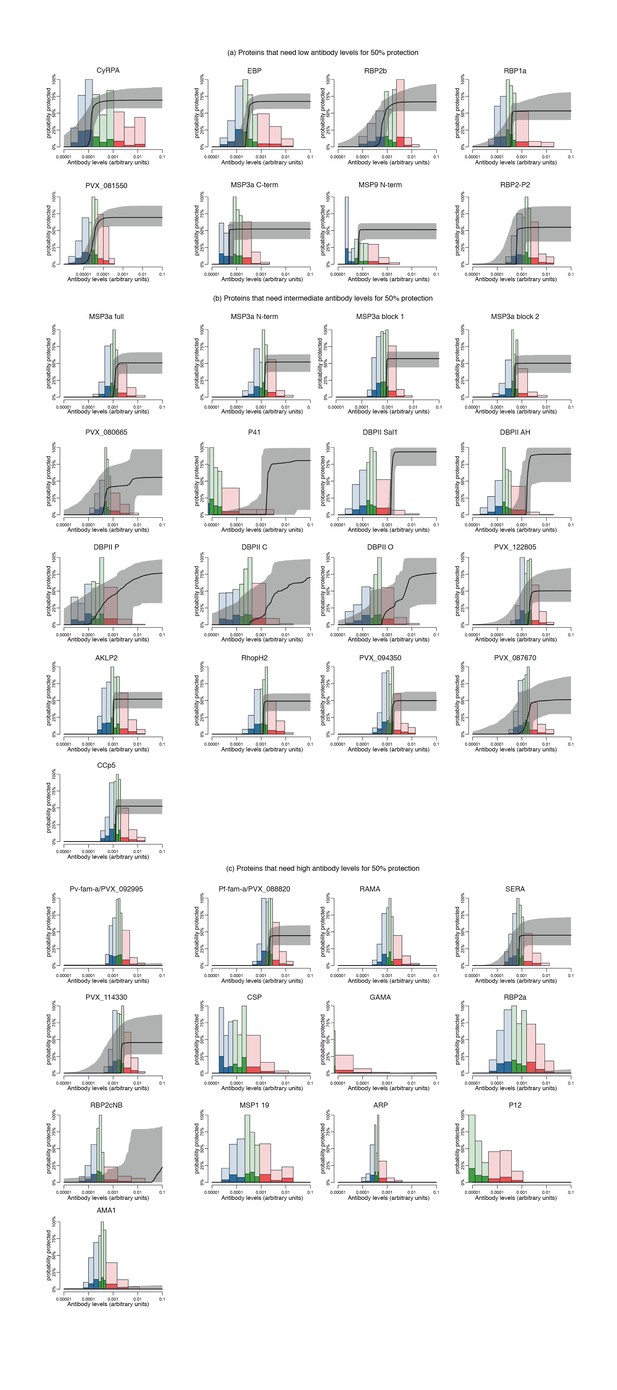
Estimated dose-response curves for the associations between antibody levels to P. vivax antigens and protection from clinical malaria.
Solid black lines depict exposure-adjusted dose-response curves, and the grey shaded regions depict the 95% credible intervals. Histograms show the observed distribution of antibody levels (arbitrary antibody units relative to standard curves made of immune pooled serum), and the darkly-colored portions of the histograms show the proportion of individuals with that antibody levels who had a P. vivax episode (>500 parasites/μL). (a) Antigens that need low antibody levels to show an association with 50% of protection against clinical malaria. (b) Antigens that need medium antibody levels to show an association with 50% of protection against clinical malaria. (c) Antigens that need high/very high antibody levels to show an association with 50% of protection against clinical malaria.
Tables
Seroprevalence of antibodies to 38 P. vivax proteins in Papua New Guinean children aged 1–3 years.
https://doi.org/10.7554/eLife.28673.002| No. of children (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Location | Protein | Geom mean* | 95% CI* | 1% of adult levels | 5% of adult levels | 10% of adult levels | 25% of adult levels | 50% of adult levels | |
| GPI-anchored merozoite surface | MSP1 19 | 0.47 | 0.38 | 0.57 | 152 (67.9) | 67 (29.9) | 38 (17.0) | 21 (9.4) | 13 (5.8) |
| P12 | 0.02 | 0.02 | 0.03 | 33 (14.7) | 2 (0.9) | 0 | 0 | 0 | |
| Peripheral surface | MSP3a full | 1.17 | 1.05 | 1.31 | 222 (99.1) | 121 (54.0) | 51 (22.8) | 12 (5.4) | 3 (1.3) |
| MSP3a block 1 | 0.79 | 0.72 | 0.86 | 222 (99.1) | 82 (36.6) | 18 (8.0) | 3 (1.3) | 0 | |
| MSP3a block 2 | 0.54 | 0.48 | 0.60 | 202 (90.2) | 48 (21.4) | 13 (5.8) | 4 (1.8) | 1 (0.4) | |
| MSP3a N-term | 0.11 | 0.10 | 0.13 | 222 (99.1) | 139 (62.0) | 64 (28.6) | 11 (4.9) | 4 (1.8) | |
| MSP3a C-term | 0.11 | 0.10 | 0.13 | 62 (27.7) | 4 (1.8) | 1 (0.4) | 0 | 0 | |
| MSP9 N-term | 0.09 | 0.08 | 0.11 | 62 (27.7) | 7 (3.1) | 3 (1.3) | 0 | 0 | |
| P41 | 0.02 | 0.02 | 0.02 | 20 (8.9) | 8 (3.6) | 3 (1.3) | 0 | 0 | |
| SERA | 0.96 | 0.87 | 1.07 | 224 (99.6) | 93 (41.4) | 38 (16.9) | 8 (3.6) | 3 (1.3) | |
| Microneme | AMA1 | 0.41 | 0.36 | 0.47 | 167 (74.6) | 34 (15.2) | 19 (8.5) | 5 (2.2) | 1 (0.4) |
| DBPII Sal1 | 0.24 | 0.21 | 0.27 | 127 (56.7) | 17 (7.6) | 9 (4.0) | 3 (1.3) | 2 (0.9) | |
| DBPII P | 0.23 | 0.19 | 0.28 | 125 (55.8) | 26 (11.6) | 12 (5.4) | 3 (1.3) | 3 (1.3) | |
| DBPII O | 0.34 | 0.28 | 0.40 | 146 (65.2) | 38 (17.0) | 13 (5.8) | 5 (2.2) | 3 (1.3) | |
| DBPII AH | 0.24 | 0.21 | 0.27 | 128 (57.1) | 15 (6.7) | 7 (3.1) | 2 (0.9) | 2 (0.9) | |
| DBPII C | 0.23 | 0.19 | 0.27 | 125 (55.8) | 26 (11.6) | 11 (4.9) | 3 (1.3) | 3 (1.3) | |
| EBP | 0.40 | 0.33 | 0.48 | 142 (63.1) | 60 (26.7) | 36 (16.0) | 14 (6.2) | 6 (2.7) | |
| GAMA | 0.01 | 0.01 | 0.01 | 5 (2.2) | 1 (0.4) | 0 | 0 | 0 | |
| CyRPA | 0.54 | 0.42 | 0.69 | 139 (61.8) | 78 (34.7) | 54 (24.0) | 40 (17.8) | 31 (13.8) | |
| Rhoptry | ARP | 0.40 | 0.37 | 0.43 | 205 (91.1) | 17 (7.6) | 6 (2.7) | 0 | 0 |
| RBP1a | 0.41 | 0.35 | 0.47 | 162 (72.3) | 39 (17.4) | 18 (8.0) | 9 (4.0) | 3 (1.3) | |
| RBP2a | 0.86 | 0.72 | 1.04 | 186 (83.0) | 102 (45.5) | 64 (28.6) | 29 (12.9) | 13 (5.8) | |
| RBP2b | 1.19 | 1.02 | 1.38 | 209 (93.3) | 130 (58.0) | 90 (40.2) | 12 (5.4) | 3 (1.3) | |
| RBP2cNB | 0.40 | 0.34 | 0.47 | 159 (71.0) | 40 (17.9) | 29 (12.9) | 13 (5.8) | 8 (3.6) | |
| RBP2-P2 | 1.68 | 1.49 | 1.91 | 224 (100.0) | 156 (69.6) | 89 (39.7) | 24 (10.7) | 13 (5.8) | |
| RhopH2 | 1.40 | 1.26 | 1.57 | 224 (99.6) | 144 (64.0) | 72 (32.0) | 18 (8.0) | 3 (1.3) | |
| RAMA | 1.44 | 1.30 | 1.61 | 225 (100.0) | 146 (64.9) | 61 (27.1) | 20 (8.9) | 7 (3.1) | |
| Pre-erythrocytic | CSP | 0.15 | 0.12 | 0.18 | 95 (42.4) | 21 (9.4) | 8 (3.6) | 2 (0.9) | 1 (0.4) |
| PVX_080665 | 0.68 | 0.61 | 0.76 | 214 (95.5) | 59 (26.3) | 28 (12.5) | 4 (1.8) | 1 (0.4) | |
| Other | PVX_081550 | 0.03 | 0.03 | 0.04 | 6 (2.7) | 0 | 0 | 0 | 0 |
| PVX_094350 | 1.44 | 1.30 | 1.59 | 225 (100.0) | 148 (65.8) | 65 (28.9) | 15 (6.7) | 6 (2.7) | |
| AKLP2 | 1.35 | 1.20 | 1.52 | 225 (100.0) | 134 (59.6) | 71 (31.6) | 17 (7.6) | 7 (3.1) | |
| PVX_087670 | 1.71 | 1.54 | 1.89 | 225 (100.0) | 160 (71.1) | 80 (35.6) | 22 (9.8) | 7 (3.1) | |
| PVX_122805 | 2.04 | 1.85 | 2.24 | 225 (100.0) | 189 (84.0) | 108 (48.0) | 24 (10.7) | 6 (2.7) | |
| CCp5 | 1.69 | 1.52 | 1.88 | 225 (100.0) | 162 (72.0) | 79 (35.1) | 20 (8.9) | 9 (4.0) | |
| PVX_114330 | 2.16 | 1.98 | 2.37 | 225 (100.0) | 198 (88.0) | 120 (53.3) | 28 (12.4) | 4 (1.8) | |
| Pv-fam-a/PVX_088820 | 2.38 | 2.17 | 2.60 | 225 (100.0) | 209 (92.9) | 127 (56.4) | 32 (14.2) | 6 (2.7) | |
| Pv-fam-a/PVX_092995 | 1.85 | 1.70 | 2.02 | 225 (100.0) | 184 (81.8) | 93 (41.3) | 18 (8.0) | 5 (2.2) | |
-
Abbreviations: No = number; Geom mean = geometric mean; 95% CI = 95% confidence interval. *IgG levels multiplied by 1000. Values are in relative antibody units interpolated from standard curves using a 5PL logistic regression model.
Additional files
-
Supplementary file 1
P. vivax antigens included in this study.
Conc = concentration; HEK293E = human embryonic kidney 293E cells; WGCF = wheat germ cell-free. *Cd4-tagged proteins. Cd4 alone was conjugated to Luminex beads (2 μg/ml per 2.5 × 10∧6 beads) and tested in all samples as a control for background reactivity.
- https://doi.org/10.7554/eLife.28673.014
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28673.015





