A synthetic biology approach to probing nucleosome symmetry
Figures
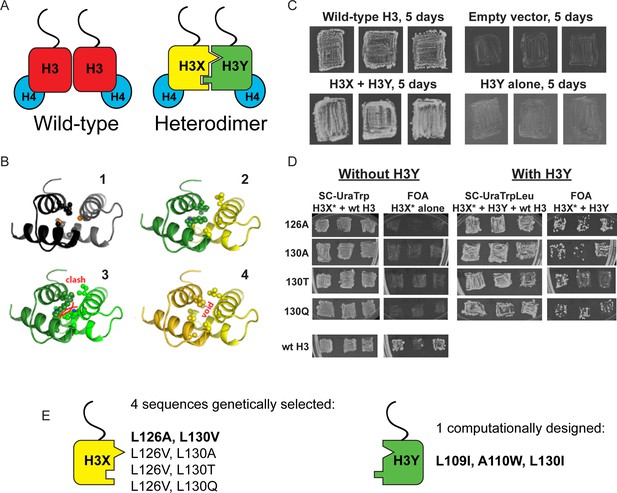
Design and testing of asymmetric nucleosomes.
(A) Schematic of asymmetric histone H3 design. Left: Wild-type H3/H4 tetramers are symmetrical, with H3-H3 interactions serving as the dimerization interface. The H3 N-terminus, site of many modifiable residues, is indicated protruding from the globular domain. Right: Obligate heterodimer H3s are comprised of distinct ‘X’ and ‘Y’ interaction partners which are altered to prevent either H3 from homodimerizing. (B) Computational models of the designed histone H3 heterodimer. (1) The four-helix bundle comprising the H3-H3 C-terminal dimerization interface (pdb 1KX3). Leucine and alanine residues at the hydrophobic interface are indicated. (2) The designed asymmetric mutations pack efficiently as a heterodimer, with partners colored in green (H3Y) and yellow (H3X). Indicated residues at the H3-H3 interface were engineered to form a bump-hole interface to increase interaction affinity. (3-4) Altered H3s are designed not to homodimerize due to van der Waals clashes (3) or voids (4) in the hydrophobic core. (C–D) Genetic analysis of heterodimeric H3X/H3Y pairs. C)H3Y alone cannot support growth. Images show growth of yeast carrying wild-type H3 on a URA3-marked plasmid as well as either empty plasmid, wild-type H3, H3Y alone, or both H3X and H3Y. Three independent transformants for each strain were grown on 5-FOA to select against the URA3 shuffle plasmid. (D) H3X alone cannot support growth. Left panels: growth of three independent transformants of various H3X strains (evolved second-generation mutations indicated at left) in the absence of H3Y. Bottom: positive control with a TRP1-marked wild-type histone plasmid. Right panels: growth of indicated H3X strains in the presence of H3Y. Synthetic complete (SC) plates were grown 3 days, FOA plates 9 days. No growth on FOA was observed for any of the four evolved H3X alleles in the absence of H3Y, even after extended incubation. (E) Sequences of final H3X-H3Y molecules. All four H3X variants have been validated genetically, while two variants – 126A/130V and 126V/130A – have been validated biochemically (see Figure 2 and not shown). 126A is used for all functional analyses.
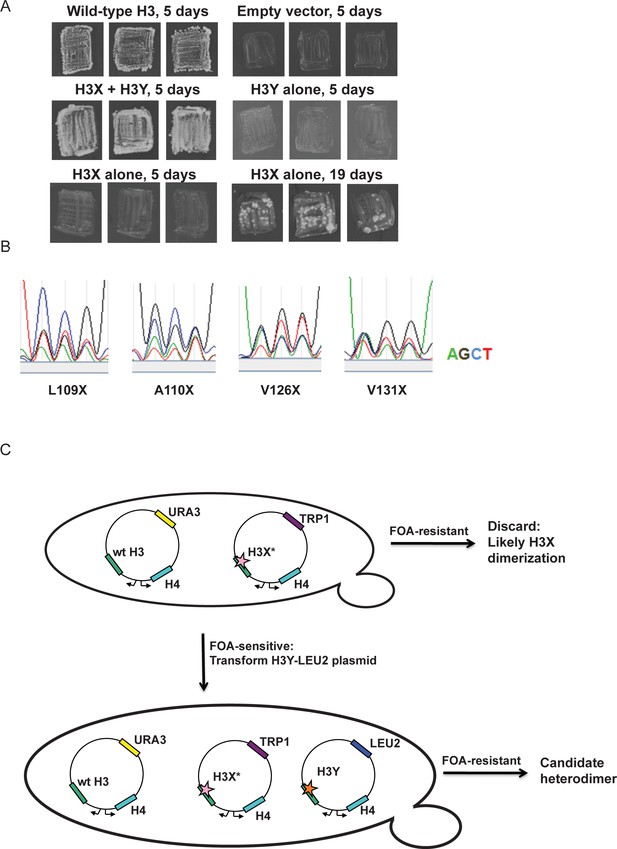
Optimization of the H3X/H3Y design.
(A) Genetic analysis of the original H3X design. The top four panels are identical to Figure 1C, showing growth of H3X/H3Y-bearing yeast strains but no growth of H3Y-bearing strains, and are duplicated here for comparison to H3X growth phenotypes. Bottom two panels: Yeast bearing a wild type histone-URA3 shuffle plasmid were transfected with the indicated plasmids carrying a wild-type H4 gene and the original H3X (126V, 130V) gene. Three independent isolates of each strain were patched onto 5-FOA media to select for loss of the wild type histone-URA3 plasmid. Strains were grown for the indicated number of days and photographed. The growth of papillae in the absence of H3Y upon extended incubation led us to optimize H3X by randomization of four key residues, as shown below. (B) Sequencing traces for the four ‘H3X*’ libraries, each with one randomly mutagenized codon as indicated. Although the base utilization in the synthetic oligonucleotides is not completely equal, all four bases are readily detected at each position. (C) Schematic of the screening strategy. Transformants carrying the H3X* library alleles were plated on SC-Trp and replicated onto FOA plates. Strains that couldn’t survive on FOA with H3* alone were transformed with an H3Y-expressing plasmid, and then were tested on FOA media to assess the viability of the H3X*-H3Y combinations (as shown in Figure 1C). As an additional criterion, we tested whether the two plasmids expressing the new H3X/H3Y heterodimers were stable when cells were cultured in rich media in the absence of selection for either plasmid. Repeated rounds of streaking on rich (YPD) plates or culturing in YPD media yielded colonies that all maintained a Trp+Leu+ phenotype, indicating that neither plasmid could be lost. Furthermore, resequencing of plasmids isolated after growth on YPD detected no reversion of the engineered H3X or H3Y in 96 independent isolates. Together, the genetic data strongly suggest that the H3X/H3Y combinations are obligate heterodimers, and that neither H3X nor H3Y can homodimerize at sufficient levels to support viability.
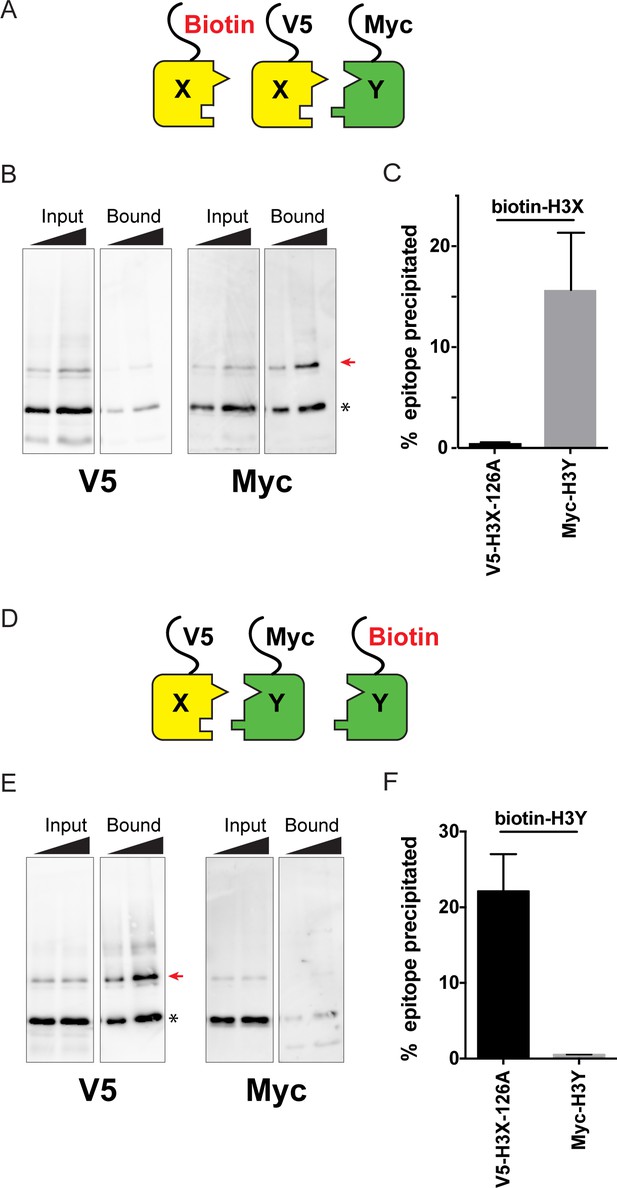
Biochemical analysis of asymmetric nucleosome formation in vivo.
(A) Schematic for in vivo biochemical analysis of H3X dimerization. Yeast strains expressed biotin-tagged H3X along with tagged H3X and H3Y, as indicated. (B) Immunoblot analysis of biotin-H3X partners. For each of the indicated antibodies (Myc or V5, the same blot was probed sequentially), the left two lanes show total DMS-crosslinked, MNase-digested chromatin (Input), and right lanes show streptavidin-precipitated biotinylated-H3 (Bound). Arrow indicates the expected size (31 kD) of crosslinked H3-H3 dimer species – note that biotin-H3 molecules crosslinked to H4 would not be recognized by the anti-epitope antibodies used for Western blot analysis. Asterisk indicates monomeric H3 molecules – presence of V5-tagged H3X (left panel) at this position in the bound fraction reflects bead contamination by uncrosslinked H3 molecules. Full gel, including unbound material and DMS control, is shown in Figure 2—figure supplement 2. (C) Quantitation of the amounts of the crosslinked 31kD H3-H3 dimers precipitated with streptavidin-agarose for second-generation H3X variant 126A. The mean and standard deviation for three independent replicate experiments are shown. (D–F) As in (A–C), but for biotin-tagged H3Y. As above, the homodimer signal (Myc, right panel) at 31kD in the bound material was almost undetectable, while V5-tagged H3X/H3Y heterodimers were readily detected. The mean and standard deviation for three independent replicate experiments are quantitated in panel (F).
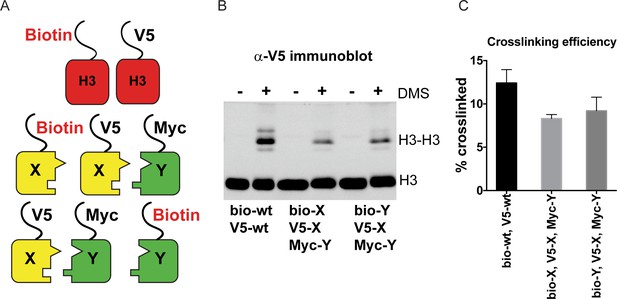
DMS crosslinking efficiency.
(A) Schematic for in vivo biochemical analysis of H3 dimerization. Yeast strains used were PKY4616 (top, biotin-tagged wild-type H3 +V5 tagged wild-type H3), PKY4625 (middle, biotin-H3X + V5-H3X + Myc-H3Y), and PKY4694 (bottom, biotin-H3Y + V5-H3X + Myc-H3Y). (B) Immunoblot analysis of strains shown in (A). Whole cell extracts after 0 or 60 min of DMS crosslinking (- DMS or +DMS, respectively) were separated by 17% SDS-PAGE, transferred to a membrane, and probed with anti-V5 antibody. (C) Quantitation of the dimer/monomer ratio of H3 proteins after DMS crosslinking. The mean and standard deviation for three independent replicate experiments are graphed.
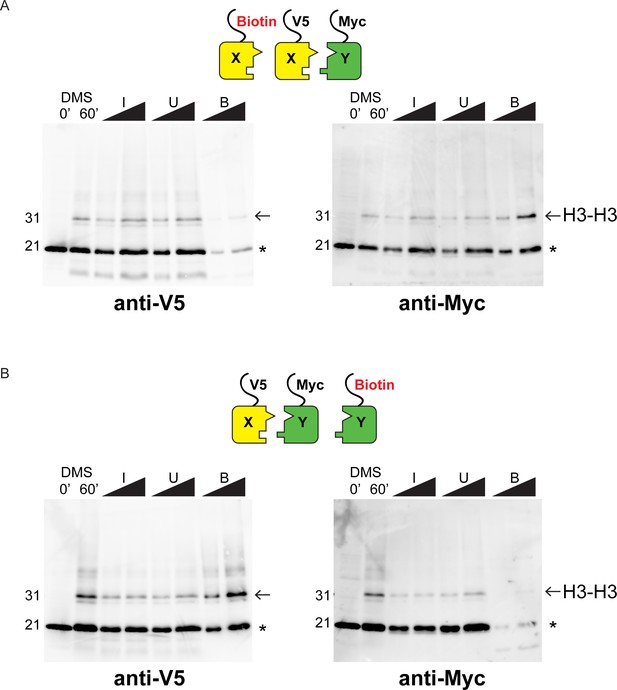
Western blot analysis of asymmetric nucleosome formation in vivo.
(A) Immunoblot analysis of Biotin-H3X126A strain. Lanes 3–4 and 7–8 from these gels are reproduced in Figure 2B,E. Lanes 1–2: whole cell extracts after 0 or 60 min of DMS crosslinking. Lanes 3–8: DMS-crosslinked, MNase-digested chromatin from the indicated strains was precipitated with streptavidin-agarose to capture biotinylated (Bio) H3. I = Input, U = Unbound: 0.75% and 1.5% of total shown in adjacent lanes; B = Bound: 10% and 20% of total shown. Proteins were separated by 17% SDS-PAGE, transferred to a membrane, and probed sequentially with the indicated primary antibodies. Arrow indicates the expected size of crosslinked H3-H3 dimer species, while asterisk indicates monomeric H3 molecules – presence of V5-tagged H3X (left panel) at this position in the bound fraction reflects bead contamination by uncrosslinked H3 molecules. (B) As in A, for biotin-H3Y.
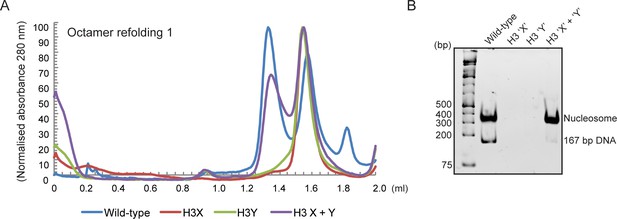
Human histone H3 mutants ‘X’ and ‘Y’ form obligate heterodimers in vitro.
(A) Size exclusion (Superdex 200 Increase 3.2/300) gel filtration profiles showing the purification of octamer refolding reactions containing wild-type and heterodimer ‘X’ and ‘Y’ histone H3. Octamer refolding in the presence of both ‘X’ and ‘Y’ H3 yields histone octamers with a characteristic elution profile, similar to refolding reactions containing wild-type H3, while reconstitutions with only H3X or only H3Y form aggregates. (B) Native PAGE analysis of nucleosome reconstitution experiments with purified histones from (A). H3X/Y octamer reconstitutions readily form nucleosomes when assembled onto DNA, while the aggregates formed in octamer refolding reactions containing either ‘X’ or ‘Y’ alone do not form histone octamers and therefore cannot form nucleosomes in vitro.
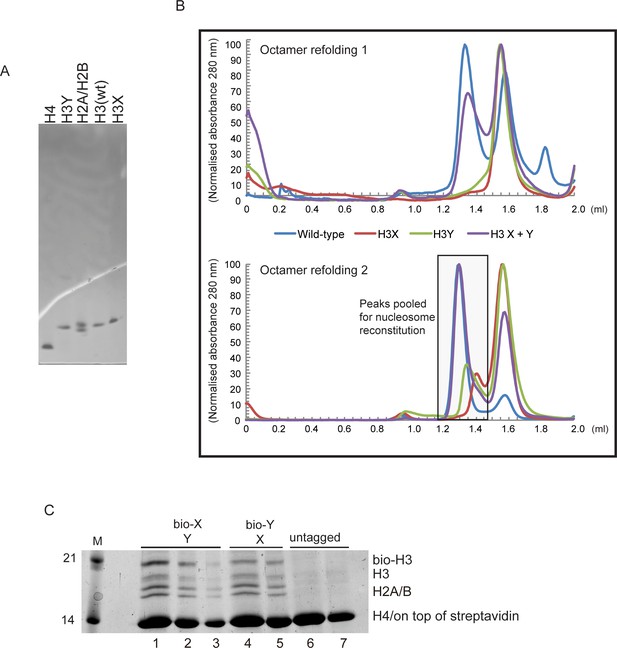
Generation of asymmetric X/Y nucleosomes for in vitro studies.
(A) Purified recombinant histones. (B) Replicate size exclusion (Superdex 200 Increase 3.2/300) gel filtration profiles showing the purification of octamer refolding reactions containing wild-type and heterodimer ‘X’ and ‘Y’ histone H3. As in Figure 3A, with additional replicate shown. (C) Coomassie Blue G staining of affinity-purified chromatin composed of asymmetric nucleosomes. Chromatin was washed with buffer containing 0.1% Triton X100 and 0.5 M NaCl and purified by binding to streptavidin-agarose in the same buffer. Strains expressed the indicated combination of X-Y asymmetric H3s. 10 μl (lanes 1, 4, 6), 5 μl (lanes 2, 5, 7) or 2.5 μl (lane 3) of the bound material was analyzed on a 15% SDS-PAGE gel.
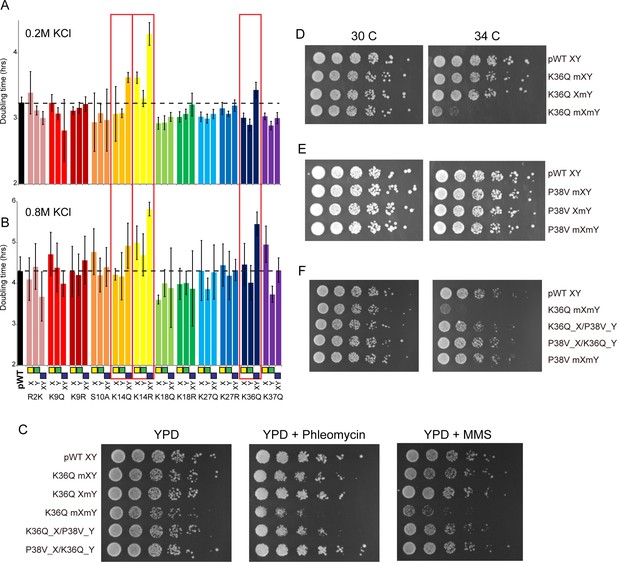
Genetic epistasis between the H3 tails.
(A–B) We constructed 12 sets of point mutations in histone H3, with each mutant cycle including H3Xmut/H3Ywt, H3Xwt/H3Ymut, and H3Xmut/H3Ymut – for each mutant trio, X/Y/double mutations are ordered from left to right, as indicated with green/yellow/blue boxes in Panel (B). For all 12 trios, as well as the matched ‘pseudo-wt’ (labeled as pWT) carrying H3Xwt/H3Ywt (leftmost bar and dashed line), we measured doubling times in at least six replicate cultures in low (A) and high (B) KCl. Boxes emphasize H3K14R and H3K36Q trios, as detailed in text. Note that growth rates here were measured robotically in a 96-well format, so doubling times are slower than those typically measured in well-aerated cultures (Figure 4—figure supplement 1A). (C) Effects of H3K36Q mutants on DNA damage sensitivity. Serial dilutions of the indicated strains were plated on YPD, YPD + 10 ng/ml phleomycin or YPD + 0.025% MMS, as indicated. pWT XY (PKY4704), K36Q mXY (PKY4829), K36Q XmY (PKY4831), K36Q mXmY (PKY4834), K36Q_X/P38V_Y (PKY5138), and P38V_X/K36Q_Y (PKY5140). (D–F) Serial dilution growth assay for the indicated strains incubated at 30 C or 34 C, as indicated. Note that yeast carrying pseudo-wild type H3X/H3Y nucleosomes grow poorly at 37C, so we assayed temperature sensitivity at 34 C. Mutant set in (D) shows temperature-sensitivity is specific to symmetric H3K36Q mutant. Mutant set in (E) shows that complete loss of H3K36me3 in the symmetric H3P38V mutant (where H3K36me3 is replaced with H3K36me2) is compatible with rapid growth at high temperatures. Mutant set in (F) shows that a single dimethylated K36 residue per nucleosome is compatible with rapid growth at high temperatures. Strains analyzed were: pWT XY (PKY4704), R2K mXY (PKY4749), R2K XmY (PKY4751), R2K mXmY (PKY4753), K9Q mXY (PKY4714), K9Q XmY (PKY4715), K9Q mXmY (PKY4706), K9R mXY (PKY4773), K9R XmY (PKY4775), K9R mXmY (PKY4777), S10A mXY (PKY4716), S10A XmY (PKY4717), S10A mXmY (PKY4707), K14Q mXY (PKY4789), K14Q XmY (PKY4791), K14Q mXmY (PKY4793), K14R mXY (PKY4781), K14R XmY (PKY4783), K14R mXmY (PKY4786), K18Q mXY (PKY4805), K18Q XmY (PKY4807), K18Q mXmY (PKY4809), K18R mXY (PKY4797), K18R XmY (PKY4799), K18R mXmY (PKY4801), K27Q mXY (PKY4822), K27Q XmY (PKY4823), K27Q mXmY (PKY4825), K27R mXY (PKY4813), K27R XmY (PKY4815), K27R mXmY (PKY4817), K36Q mXY (PKY4829), K36Q XmY (PKY4831), K36Q mXmY (PKY4834), K37Q mXY (PKY4837), K37Q XmY (PKY4839), K37Q mXmY (PKY4841), P38V mXY (PKY5033), P38V XmY (PKY5035), P38V mXmY (PKY5037), K36Q mX/P38V mY (PKY5138), and P38V mX/K36Q mY (PKY5140). pWT XY, pseudo WT; mXY, mutation on X; XmY, mutation on Y; and mXmY, mutation on both X and Y.
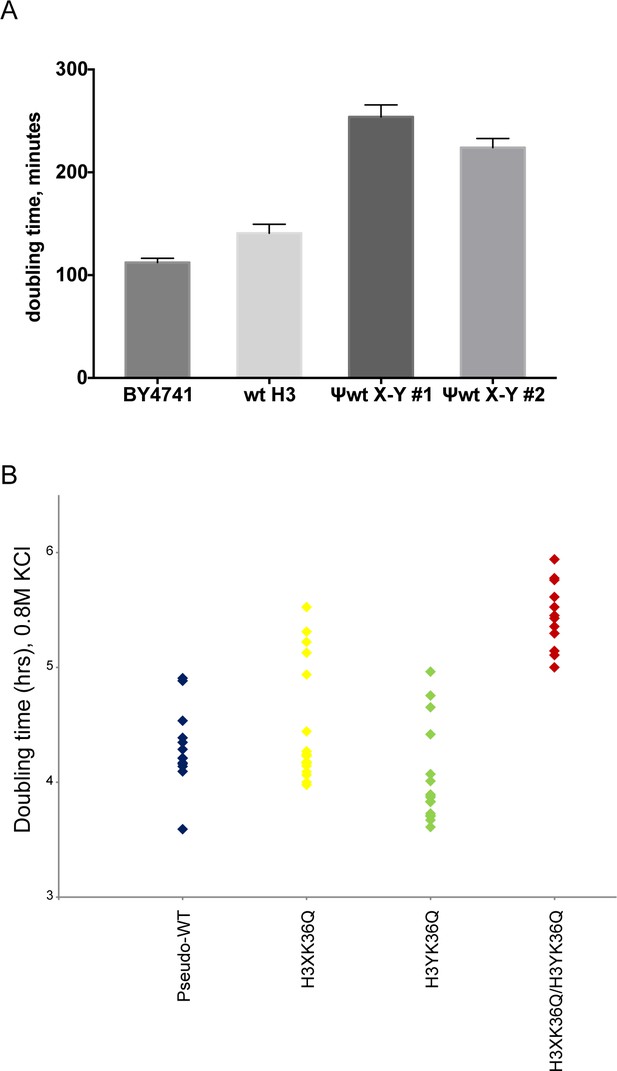
Significant growth defects for the H3K36Q double mutant, relative to either single mutant.
(A) The X/Y interface confers slow growth relative to the wild-type H3 interface. Growth rates of the indicated strains are shown for growth in liquid YPD media at 30C in well-aerated flasks. Strains analyzed were BY4741 (wild-type strain with no histone gene deletions), ‘wt H3’=PKY4701, the histone shuffle strain with a plasmid expressing wild-type H3/H4, ψwt X-Y#1 and #2 = PKY4704 and 4705, two different PKY4701 derivatives coexpressing the heterodimeric X-Y H3 derivatives. (B) Dot plots show all growth rates in 0.8M KCl for all individual replicate time courses for the indicated H3K36Q mutants. These measurements were performed in 96-well plates.
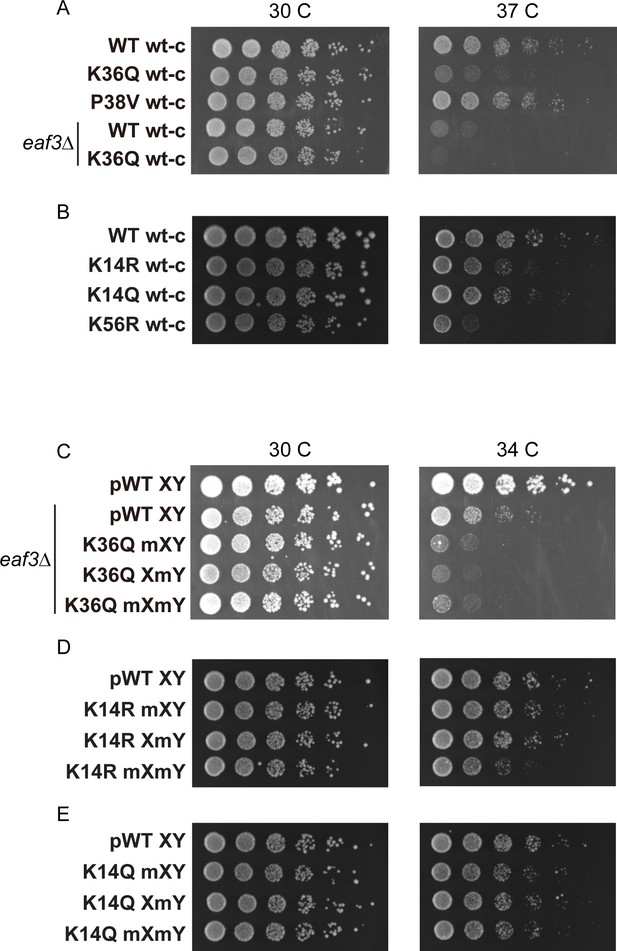
Temperature sensitivity of various histone mutant strains.
For all panels, serial dilutions of the indicated strains were plated at 30 C, 34 C, or 37 C, as indicated. (A) Data for WT wt-c (PKY4701), K36Q wt-c (PKY4827), P38V wt-c (PKY5031), eaf3Δ WT wt-c (PKY5077) and eaf3Δ K36Q wt-c (PKY5079). (B) Data for WT wt-c (PKY4701), K14R wt-c (PKY4779), K14Q wt-c (PKY4787) and K56R wt-c (PKY4843). (C) Data for pWT XY (PKY4704), eaf3Δ pWT XY (PKY5081), eaf3Δ K36Q mXY (PKY5083), eaf3Δ K36Q XmY (PKY5086) and eaf3Δ K36Q mXmY (PKY5087). (D) Data for pWT XY (PKY4704), K14R mXY (PKY4781), K14R XmY (PKY4783) and K14R mXmY (PKY4786). (E) Data for pWT XY (PKY4704), K14Q mXY (PKY4789), K14Q XmY (PKY4791) and K14Q mXmY (PKY4793) pWT_XY, pseudo WT; mXY, mutation on X; XmY, mutation on Y; and mXmY, mutation on both X and Y.
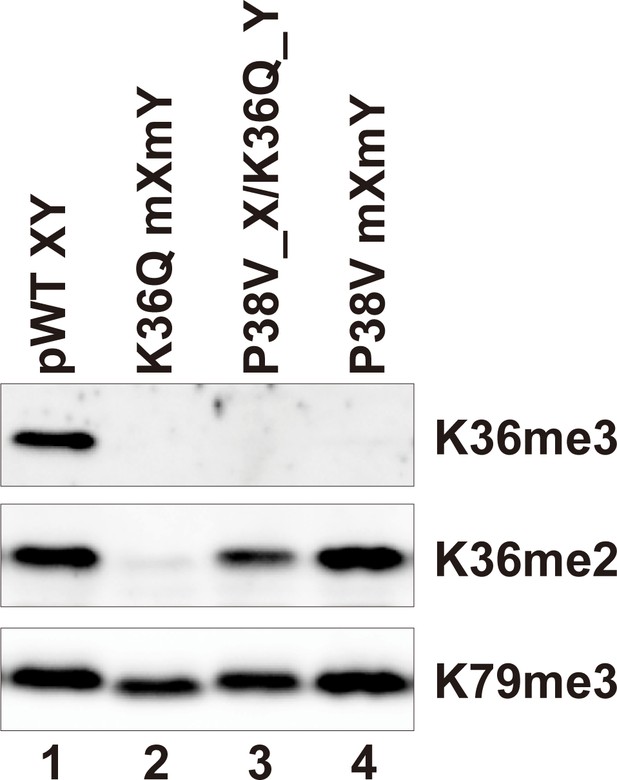
H3P38V mutations block trimethylation but not dimethylation of H3K36.
Whole cell extracts of the indicated strains were separated by 15% SDS-PAGE and transferred to a membrane. The same blot was probed sequentially with the following antibodies: anti-H3K36me3 (Active Motif MABI0333), anti-H3K36me2 (Abcam ab9049) and anti-H3K79me3 (GeneTexRM157). Strains analyzed were: pWT XY (PKY4704), K36Q mXmY (PKY4834), P38V_X/K36Q_Y (PKY5140), P38V mXmY (PKY5037).
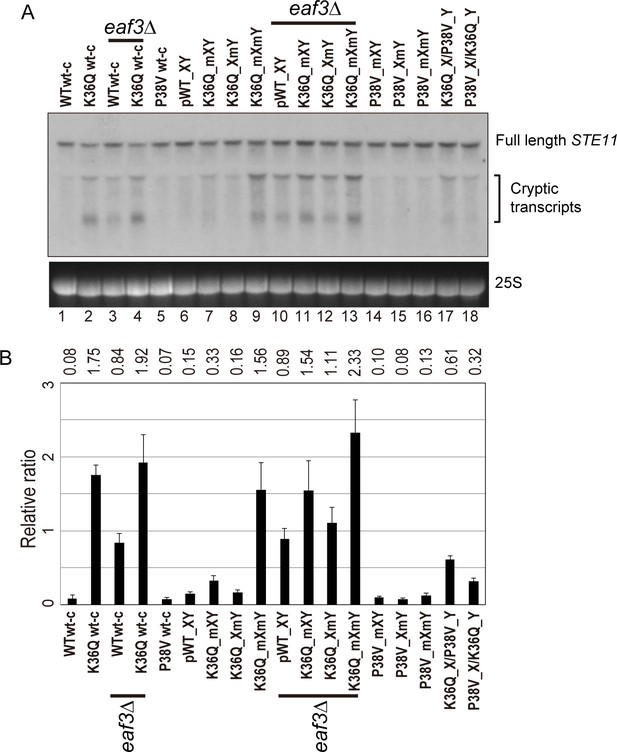
Double H3K36Q mutants fail to repress cryptic promoters.
(A) Northern blot for STE11 for RNA isolated from the indicated strains, with 25S Northern shown as a loading control. Top band, which migrates at 2.2 kb, represents the full-length STE11 transcript, while the two lower molecular-weight bands correspond to previously-characterized sense transcripts initiating within the STE11 coding region from cryptic promoters (Kaplan et al., 2003). (B) Quantitation of cryptic transcript levels for STE11 in the various strains indicated (n = 3). Levels of cryptic transcripts were normalized to full-length. Average and std. dev. of triplicate measures of these normalized values are graphed on the y-axes. Strains analyzed were: WT wt-c (PKY4701), K36Q wt-c (PKY4827), eaf3Δ WT wt-c (PKY5077), eaf3Δ K36Q wt-c (PKY5079), P38V wt-c (PKY5031), pWT XY (PKY4704), K36Q mXY (PKY4829), K36Q XmY (PKY4831), K36Q mXmY (PKY4834), eaf3Δ pWT XY (PKY5081), eaf3Δ K36Q mXY (PKY5083), eaf3Δ K36Q XmY (PKY5086) and eaf3Δ K36Q mXmY (PKY5087), P38V mXY (PKY5033), P38V XmY (PKY5035), P38V mXmY (PKY5037), K36Q mX/P38V mY (PKY5138), and P38V mX/K36Q mY (PKY5140). pWT_XY, pseudo wild-type; mXY, mutation on X; XmY, mutation on Y; and mXmY, mutation on both X and Y.
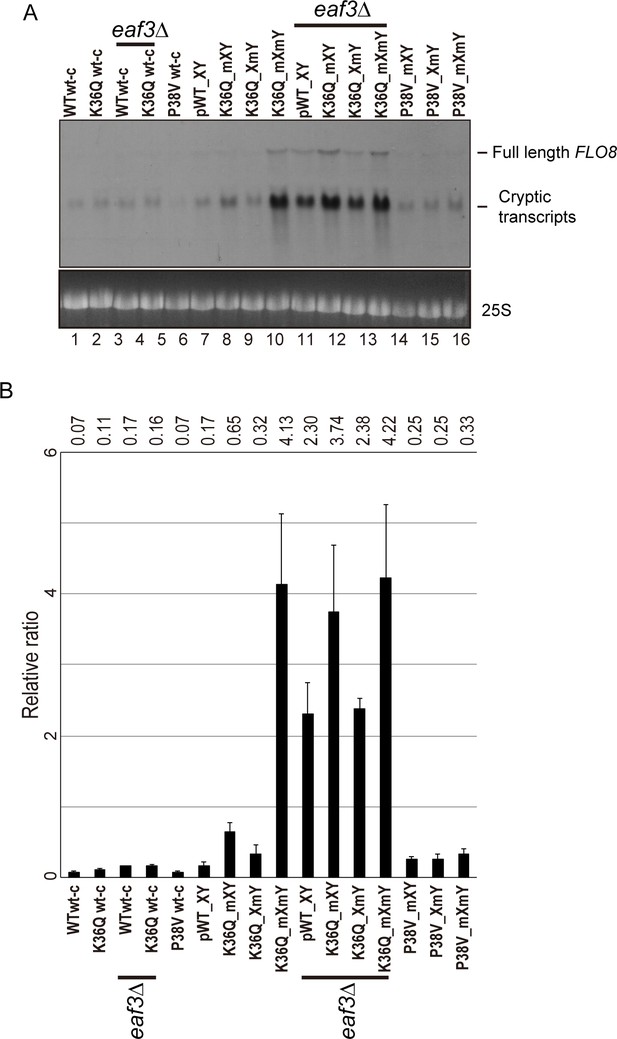
Effects of H3K36Q mutants on cryptic transcription.
As in Figure 5, with FLO8 used as a probe for Northern blots in place of STE11.
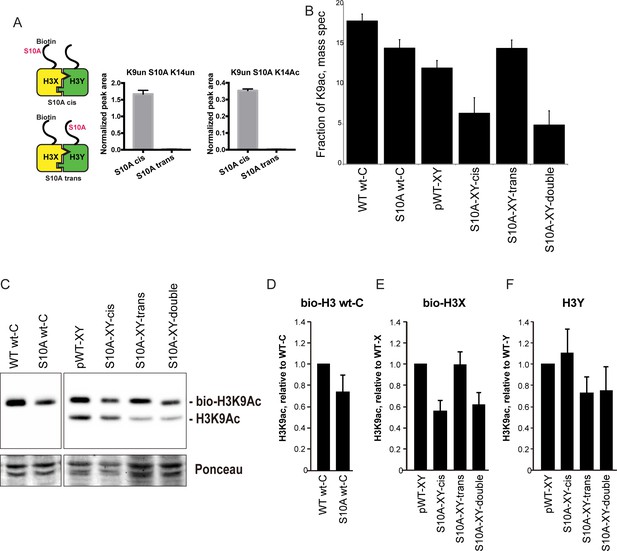
H3S10A affects histone crosstalk in cis.
(A) Mass spectrometric analysis of H3X/H3Y heterodimers expressing one biotin-labeled subunit, with a S10A point mutation either located on the same H3 molecule (in cis), or in trans. Left: Schematic of asymmetric nucleosomes. Right: Robust avidin-affinity purification of biotin-labeled H3 molecules. Mass spec quantitation of peptides (described on top of the graphs) from the strains with genotypes indicated on the x-axes. For each peptide, raw peak area scores were normalized by dividing by the peak areas of an internal control, the unmodified H3 peptide containing K42. Average and std. dev. of triplicate measures of these normalized values are graphed on the y-axes. As expected, S10A-containing peptides were only detected when present in cis on the biotin-labeled histone, but not when expressed in trans on the heterodimeric partner. (B) H3K9 acetylation levels are diminished in cis in H3S10A mutants. Data are shown for mass spec analysis of the indicated mutant strains. Left two bars show data for wild-type and double H3S10A mutants on the background of a wild-type H3-H3 interface. Right four panels show H3K9Ac levels on an avidin-purified biotin-tagged H3 molecule, showing data for H3X acetylation in the context of the pseudo-wild type H3X/Y background, H3X acetylation on the same (cis) or opposite tail from an H3S10A mutation, and H3X acetylation in a double H3XS10A/H3YS10A mutant. (C) Western blots confirming effects of symmetric S10A on K9 acetylation both on the WT H3-H3 background, and on the X-Y background, as indicated. For the right panel, the epitope tag present on H3X allows separate probing of the H3X and H3Y molecules. Total protein on blots was visualized by Ponceau S staining. Note that although the S10A mutation in principle might affect anti-H3K9ac antibody (Abcam ab10812) binding, these results are highly concordant with the mass spectrometry measurements in panel (B), where the ability to detect lysine acetylation (based on peptide mass) is unaffected by the difference between S10 and S10A. (D–F) Quantitation of blots from panel (C). K9Ac signals were normalized to total protein detected by Ponceau S. Strains analyzed were: WT wt-C (PKY4610), S10A wt-C (PKY5003), pWT-XY (PKY4983), S10A-XY-cis (PKY5005), S10A-XY trans (PKY4986) and S10A-XY double (PKY5042).
Additional files
-
Supplementary table 1
Yeast strain list.
- https://doi.org/10.7554/eLife.28836.016
-
Transparent reporting form
- https://doi.org/10.7554/eLife.28836.017





