Evolutionary transitions between beneficial and phytopathogenic Rhodococcus challenge disease management
Figures
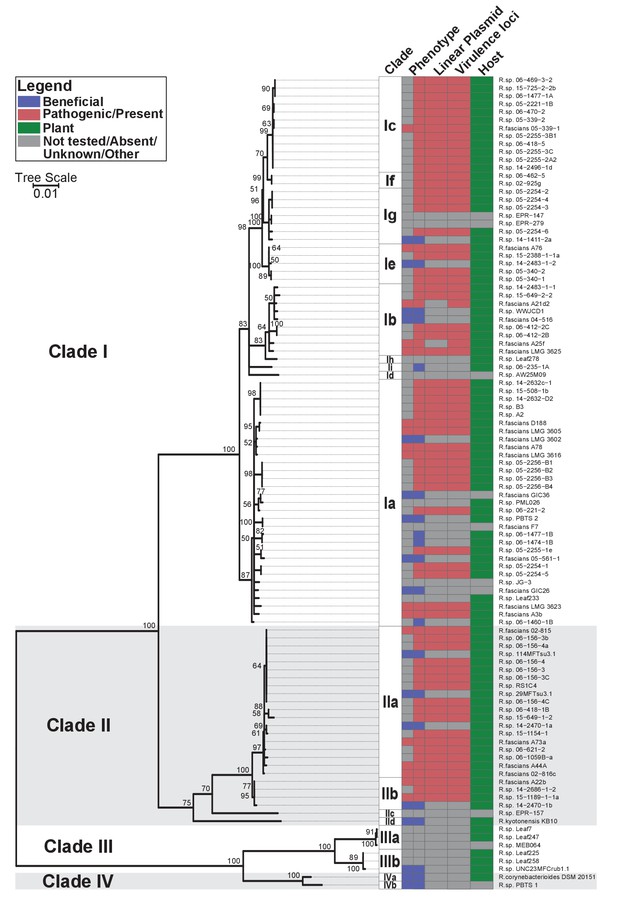
Plant-associated isolates of Rhodococcus form four sister clades.
Multi-locus sequence analysis maximum likelihood tree of plant-associated isolates of Rhodococcus. Translated sequences for ftsY, infB, rpoB, rsmA, secY, tsaD, and ychF from 104 members of Rhodococcus were identified using TBLASTN, aligned, and used to generate a multi-locus maximum likelihood tree. Clade designations are based on analysis of average nucleotide identity (Supplementary file 1B). Columns indicate the features of the corresponding isolate. Grey bars indicate not tested, absent, unknown, or other for phenotype, linear plasmid, virulence loci, and host columns, respectively. The left-half of the column corresponding to phenotype indicates a confirmed phenotype, whereas the right-half indicates an inference based on the presence or absence of virulence genes.
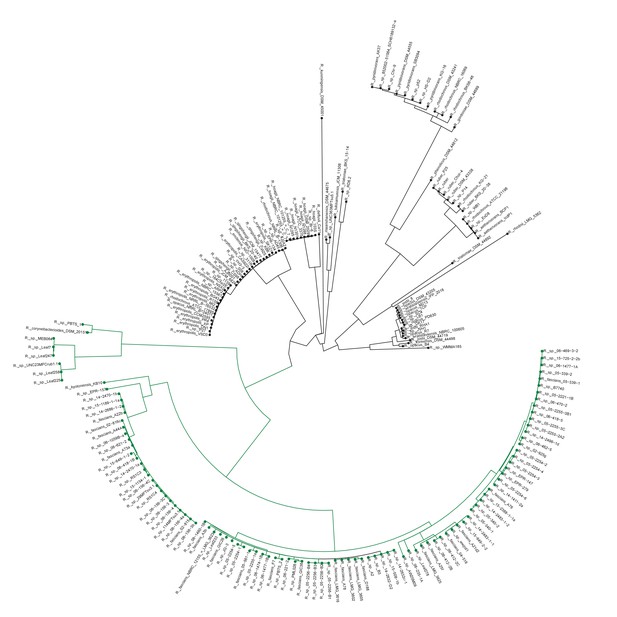
Plant-associated Rhodococcus form a distinct clade.
Multi-locus sequence analysis maximum likelihood tree of isolates of the Rhodococcus genus. Translated sequences for ftsY, infB, rpoB, rsmA, secY, tsaD, and ychF from 199 members of Rhodococcus were identified using TBLASTN, aligned, and used to generate a multi-locus maximum likelihood tree.
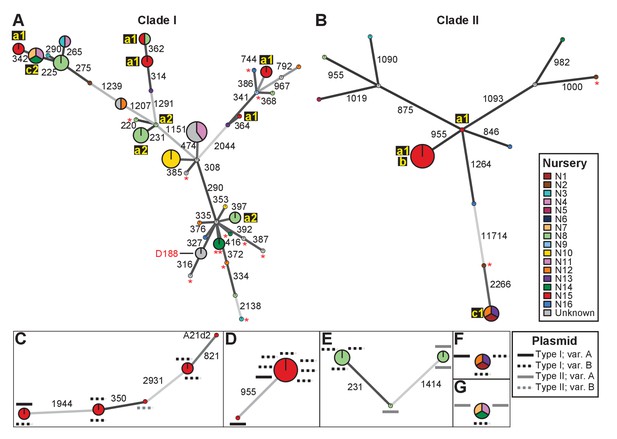
Analysis of SNPs reveals three transmission patterns of pathogenic Rhodococcus.
Minimum spanning networks of isolates of (A) Clade I and (B) Clade II. Each genotype is displayed as a circle, with sizes scaled to represent the number of associated isolates (smallest = 1 isolate). Colors represent the source of the isolates (see key), with coloring proportional to the ratio of isolates from each source. Lower-case letters and numbers (a1, a2, b, c1, and c2) highlight potential transmission patterns; see panels C–G). Asterisks = lacking virulence genes. The genotype that includes D188 is indicated. (C, D) Minimum spanning networks of pathogenic isolates belonging to Clade I (C; ‘a1’) and Clade II (D; ‘a1’ and ‘b’) from nursery N15. A21d2 lacks a virulence plasmid and its virulence loci are present in the chromosome. (E) The minimum spanning network for isolates of pathogenic isolates from nursery N8 (‘a2’). (F) The epidemiological link ‘c1’ between isolates from nurseries N1, N12, and N13. (G) The epidemiological link ‘c2’ between isolates from nurseries N7, N11, and N14. Plasmid types and their variants are mapped onto each of the nodes (see key). Numbers adjacent to connecting lines indicate the number of SNPs that separate each genotype. The lengths of connecting lines are arbitrary; gray lines indicate distances that exceed an arbitrary threshold.
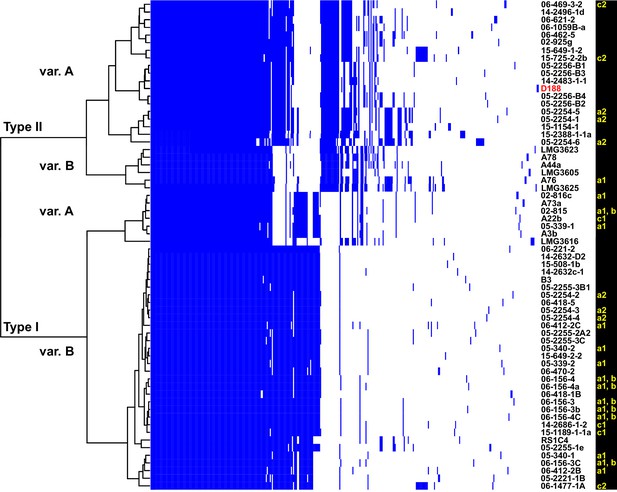
Analysis of plasmid variation reveals multiple patterns in distribution.
Rows indicate genes present in (blue) or absent from (white) the plasmids of isolates (listed to the right; D188 is labeled in red for reference). Columns represent individual genes. Type categories were determined on the basis of phylogenetic analysis of the core genes. INDEL variants, delineated by gray and white shading, were determined on the basis of the cladogram. The plasmid carried by LMG3616 is enclosed by dotted lines. The lower-case letters and numbers (a1, a2, b, c1, and c2) listed along the right, relate isolates and their plasmids to the potential transmission patterns indicated in Figure 2.
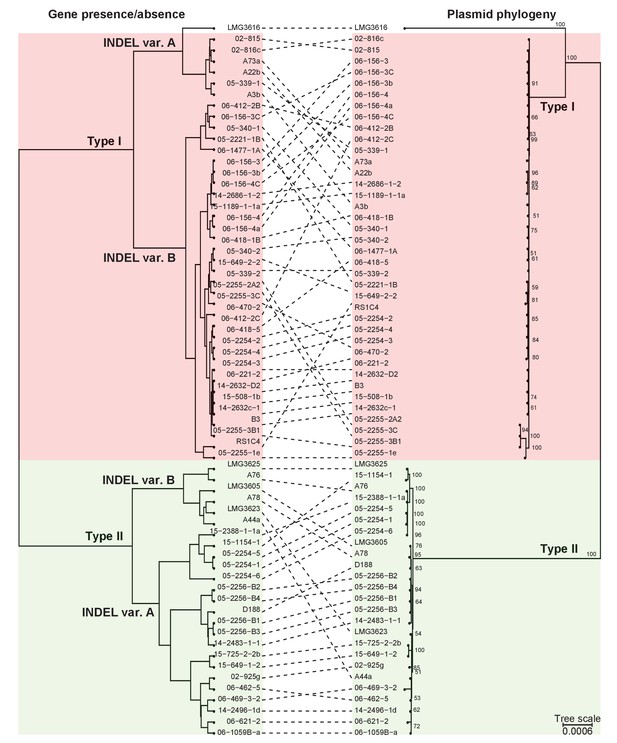
Presence/absence-cladogram and phylogeny support the classification scheme for the virulence plasmid.
The dashed lines show congruency in results from methods used to cluster plasmid types; (left) plasmids were clustered in a cladogram on the basis of the presence/absence of genes and (right) in a phylogenetic tree on the basis of the concatenated sequences of 123 genes that are present in more than 95% of the virulence plasmid sequences. Red and green blocks delineate Type I and Type II plasmids, respectively.
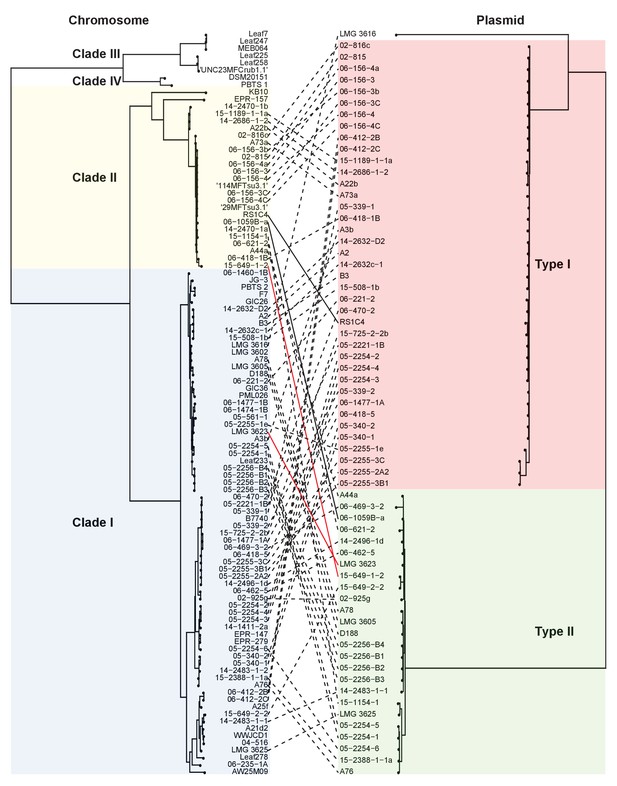
Both types of virulence plasmids are present in both clades of pathogenic Rhodococcus.
The multilocus sequence analysis (MLSA) phylogenetic tree of the four plant-associated clades was associated with the phylogenetic tree on the basis of the concatenated sequences of 123 genes that are present in more than 95% of the virulence plasmid sequences. The lines between trees show relationships between isolates and plasmids. The two pairs of solid (black and red) lines highlight examples of closely related isolates that have divergent plasmid types and distantly- related isolates that have similar plasmid types.
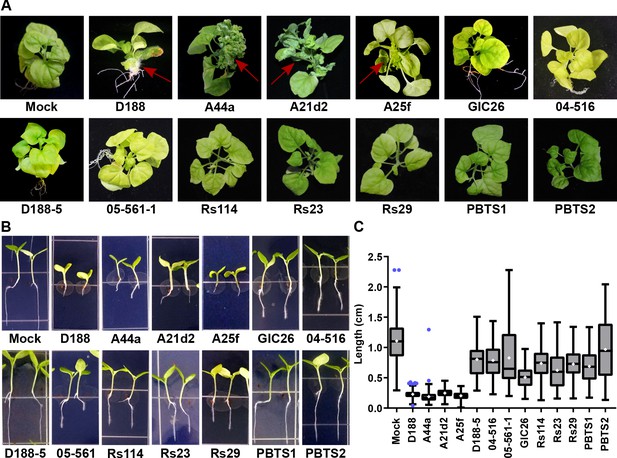
The att, fasR, and fas loci are necessary for the pathogenicity of Rhodococcus.
(A) Representative images of leafy galls on N. benthamiana. Isolates of Rhodococcus were inoculated at the apical meristem. Red arrows indicate the leafy galls. Rs114, Rs23, and Rs29 are abbreviations for isolates 114MFTsu3.1, UNC23MFCrub1.1, and 29MFTsu3.1, respectively. (B) Representative images of the root length of seedlings. Three-day-old N. benthamiana seedlings were inoculated with theindicated isolate of Rhodococcus or water (mock) and grown vertically for seven days under constant light. Isolates D188, A44a, A21d2, and A25f are the only isolates with virulence genes .(C) Quantification of seedling root length. All treatments, except for PBTS2, were significant compared to the mock treatment.
-
Figure 4—source data 1
Lengths of N. benthamiana seedling roots 7 days after inoculation with wild type Rhodococcusisolates.
- https://doi.org/10.7554/eLife.30925.010
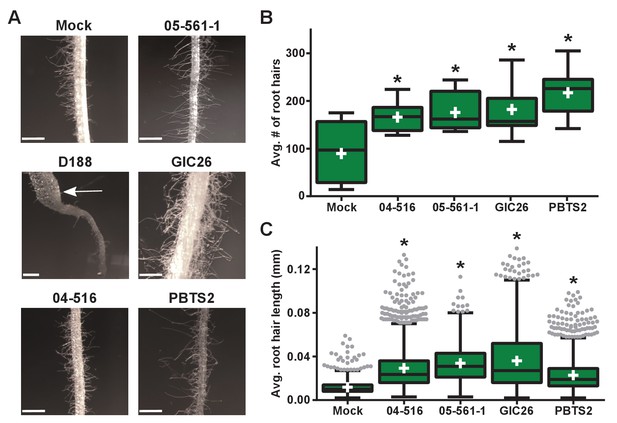
Plant-associated Rhodococcus bacteria cause changes to the root architecture of seedlings.
(A) Representative images of root hairs of N. benthamiana that were inoculated with isolates of Rhodococcus. Images were taken 25 days post inoculation (dpi). The white arrow indicates the thicker stem induced only by isolate D188. Scale bars are 0.5 mm. (B) Quantification of average root hair number at 25 dpi. All root hairs were manually counted for at least five seedlings per treatment. (C) Quantification of root hair lengths at 25 dpi. All root hairs were manually measured for at least five seedlings per treatment. For B and C, data were repeated in two independent biological replicates. * indicates a significant difference compared to the mock treatment.
-
Figure 5—source data 1
Numbers of N. benthamiana seedling root hairs 25 days after inoculation with wild type Rhodococcus isolates.
- https://doi.org/10.7554/eLife.30925.012
-
Figure 5—source data 2
Lengths of N. benthamianaroot hairs 25 days after inoculation with wild type Rhodococcus isolates.
- https://doi.org/10.7554/eLife.30925.013

Five additional virulence-gene-lacking isolates of Rhodococcus cause changes to root architecture.
Representative images of seedlings inoculated with the indicated isolates of Rhodococcus or water (mock). The root lengths of seedlings inoculated with isolates lacking virulence genes were different from those of seedlings inoculated with isolate D188.
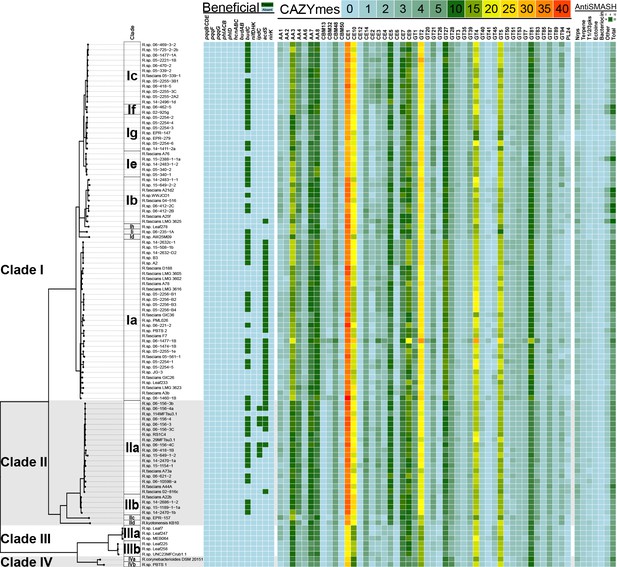
Heatmap of genes and functions enriched in clades of plant-associated Rhodococcus.
Genes associated with plant-beneficial functions were used as queries in TBLASTN searches for homologs in Rhodococcus (green = presence [>40% identity, >70% length]; cyan = absence). For the CAZYmes, only the 48 most highly represented classes are shown. The key shows the number of homologs detected. Putative secondary metabolite biosynthetic clusters were identified using antiSMASH. The key shows the number of loci predicted for each category.
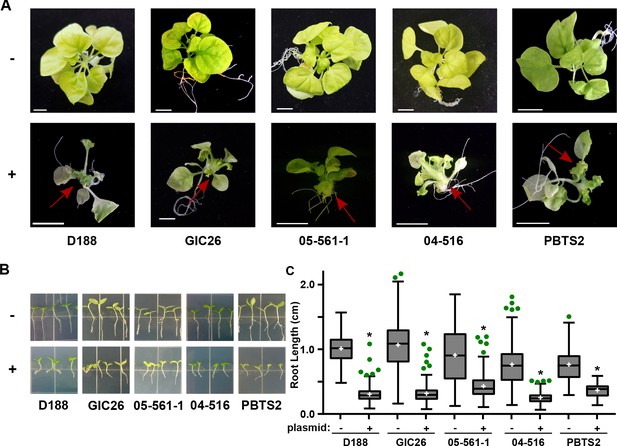
Plasmid pFiD188 with functional fasR and fas is sufficient to transition Rhodococcus isolates to phytopathogens.
(A) Representative images of leafy galls on N. benthamiana. Red arrows indicate leafy galls. Images for GIC26, 04–516, and PBTS2 are repeated from Figure 1. (B) Representative images of the root lengths of seedlings. Three-day-old N. benthamiana seedlings were inoculated with the indicated isolate of Rhodococcus or water (mock) and grown vertically for seven days under constant light. (C) Quantification of the root lengths of N. benthamiana seedlings. In all panels, -/+ indicates absence or presence of pFiD188Δatt. * indicates a significant difference compared to plants treated with the corresponding genotype lacking the plasmid.
-
Figure 6—source data 1
Lengths of N. benthamiana seedling roots 7 days after inoculation with Rhodococcusisolates +/- pFiD188Δatt.
- https://doi.org/10.7554/eLife.30925.022
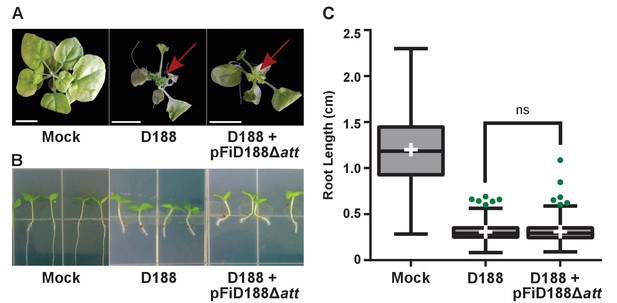
Rhodococcus isolate D188 with plasmid pFiD188Δatt is pathogenic.
(A) Representative images of leafy galls on N. benthamiana. Images were taken 56 days post inoculation (dpi). Red arrow indicates leafy gall. (B) Representative images of root length of seedlings. Three-day-old N. benthamiana seedlings were inoculated with the indicated isolate of Rhodococcus or water (mock) and grown vertically for seven days under constant light. (C) Quantification of root lengths of N. benthamiana seedlings. Data for D188 +pFiD188Δatt are the same as those shown in Figure 6.
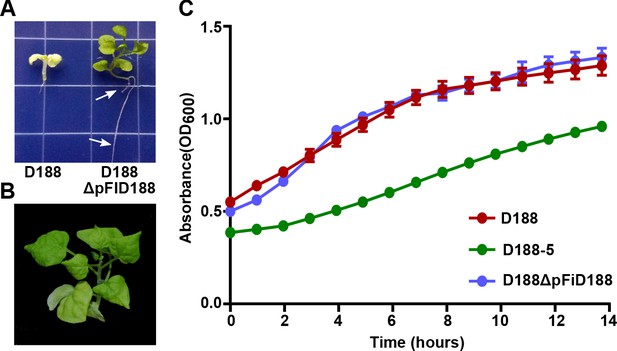
Eviction of the virulence plasmid reverts isolate D188 to a beneficial bacterium.
(A) Rhodococcus D188ΔpFID188 causes changes in root architecture. White arrows indicate lateral roots and root hairs. The image was taken 42 days post inoculation (dpi); effects were seen earlier. (B) Rhodococcus D188ΔpFID188 is not pathogenic. The strain was inoculated onto N. benthamiana. The image was taken 56 dpi. (C) Rhodococcus isolate D188-5 is compromised in in vitro growth. Rhodococcus isolates D188, D188-5, and D188ΔpFiD188 were grown separately in LB media. Growth, determined using a Tecan plate reader, was quantified on the basis of OD600 readings for a period of 16 hr. One representative experiment of three is shown; all had similar results. Error bars indicate SEM.
-
Figure 6—figure supplement 2—source data 1
Optical density values of culture-grown bacteria.
- https://doi.org/10.7554/eLife.30925.019
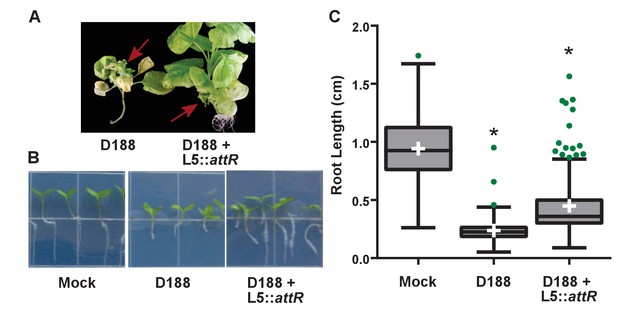
Rhodococcus isolate D188 carrying L5::attR is affected in virulence.
(A) Representative images of leafy galls on N. benthamiana. Images were taken 56 days post inoculation (dpi). Red arrows indicate a leafy gall. (B) Representative images of the root lengths of seedlings. Three-day-old N. benthamiana seedlings were inoculated with the indicated isolate of Rhodococcus or water (mock) and grown vertically for seven days under constant light. (C) Quantification of root lengths of N. benthamiana seedlings. * indicates a significant difference compared to treatment with D188.
-
Figure 6—figure supplement 3—source data 1
Lengths of N. benthamiana seedling roots 7 days after inoculation with isolate D188 +/- L5::attR.
- https://doi.org/10.7554/eLife.30925.021
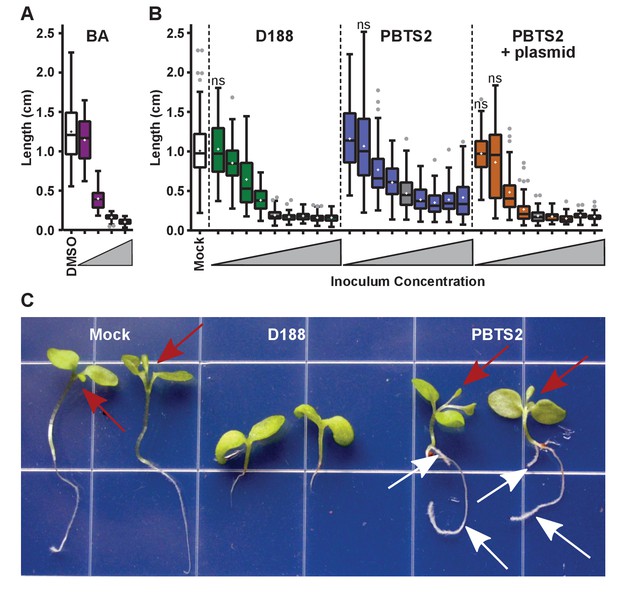
Rhodococcus has a dose-dependent effect on the root elongation of N. benthamiana seedlings.
(A) Quantification of the seedling root lengths of plants grown in exogenously applied cytokinin (6-benyzlaminopurine; BA). Three-day-old N. benthamiana seedlings were transferred to media supplemented with BA (0.01, 0.1, 1.0, and 10 µM) or dimethyl sulfoxide (DMSO). (B) Quantification of the root lengths of seedlings inoculated with increasing doses of Rhodococcus. Three-day-old N. benthamiana seedlings were inoculated with isolates D188, PBTS2, or PBTS2 + pFiD188Δatt, with doses ranging from 2.5 × 102 to 1.0 × 1012 colony-forming units (cfu). The sample shaded in gray highlights the inoculum of OD600 = 0.5 (1x = 2.5 × 1010 cfu) used in all other assays. Inocula below this decrease in 100-fold intervals. Inocula above increase at 2x, 4x, 10x, and 20x. All treatments are significantly different from mock unless otherwise noted with ‘ns’. (C) Representative image of morphological changes in seedlings. Seedlings inoculated with Rhodococcus D188 or PBTS2 (5 × 1011 cfu; 10x typical amount) or water (mock) were photographed. Red arrows indicate true leaves. White arrows indicate lateral roots and the proliferation of root hairs.
-
Figure 7—source data 1
- https://doi.org/10.7554/eLife.30925.024
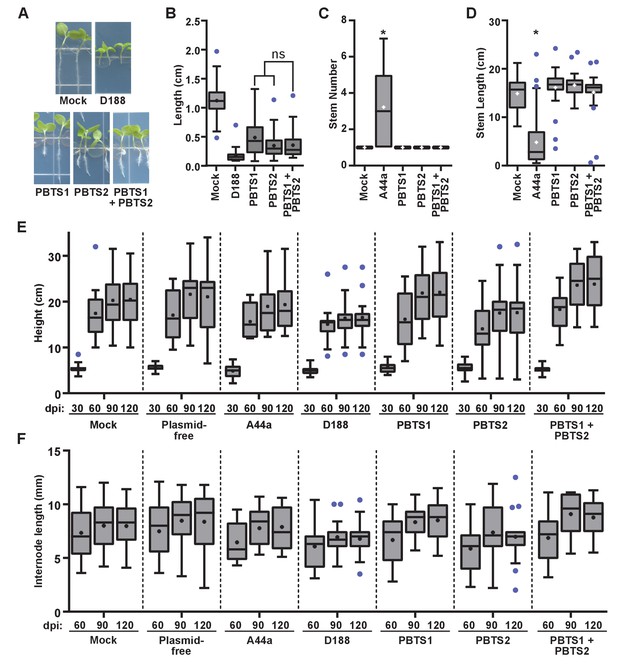
Rhodococcus isolates PBTS1 and PBTS2 do not cause disease symptoms in tested plant species.
(A) Representative images of disease symptoms or beneficial effects on seedlings of N. benthamiana. Three-day-old seedlings were inoculated with Rhodococcus or water (mock) and grown vertically for seven days under constant light. (B) Quantification of seedling root length of N. benthamiana. All were significant relative to mock. There were no significant (ns) differences between treatments with PBTS1, PBTS2, or their mixture. (C, D) Quantification of stem number (C) and stem length (D) of peas inoculated with Rhodococcus or mock-inoculated. * indicates a significant difference when comparing to the mock treatment. (E, F) Quantification of height (E) and internode length (F) of pistachio UCB1 rootstock inoculated with Rhodococcus or mock-inoculated. No treatment effect was significantly different from the time-matched mock treatment. One representative experiment of three is shown.
-
Figure 7—figure supplement 1—source data 1
Lengths of N. benthamiana seedling roots 7 days after growth in BA or inoculation with isolates D188 or PBTS2.
- https://doi.org/10.7554/eLife.30925.026
-
Figure 7—figure supplement 1—source data 2
Number of stems of peas 14 days after inoculation with isolates D188, PBTS1, or PBTS2.
- https://doi.org/10.7554/eLife.30925.027
-
Figure 7—figure supplement 1—source data 3
Length of stems of peas 14 days after inoculation with isolates D188, PBTS1, or PBTS2.
- https://doi.org/10.7554/eLife.30925.028
-
Figure 7—figure supplement 1—source data 4
Heights of pistachio UCB1 210 days after inoculations with Rhodococcus isolates.
- https://doi.org/10.7554/eLife.30925.029
-
Figure 7—figure supplement 1—source data 5
Internode lengths of pistachio UCB1 210 days after inoculations with Rhodococcus isolates.
- https://doi.org/10.7554/eLife.30925.030
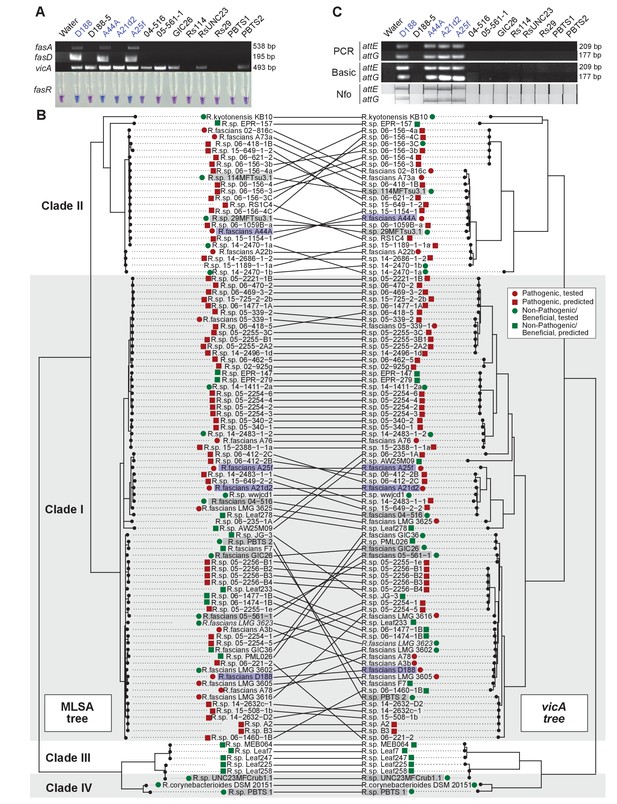
Use of the vicA gene does not discriminate pathogenic Rhodococcus.
(A) (Top) Using locus-specific primers and DNA from the listed isolates, fragments of fasA, fasD and vicA were PCR amplified. Pathogenic isolates are labeled in blue; virulence-gene-lacking isolates are labeled in black. Products were resolved on a 1% TAE agarose gel. Amplicon sizes are listed to the right of the gel images. (Bottom) LAMP assay detection of fasR from the same DNA samples. A positive result is visualized by a blue color. Negative results are light purple. (B) Congruency between species (left) and vicA trees (right). Clades other than I–IV are not shown. Highlighted isolates are the same as in A, pathogenic isolates are highlighted in blue, virulence-gene-lacking isolates are highlighted in gray. (C) Standard endpoint PCR, RPA Basic, and RPA nfo were used to detect attE or attG from DNA extracted from isolates of Rhodococcus. Pathogenic isolates are labeled in blue. For PCR and RPA basic, product sizes are list to the right of the figure. For RPA nfo, the presence of the test band is indicative of a positive reaction; the control bands for all strips were confirmed (not shown).
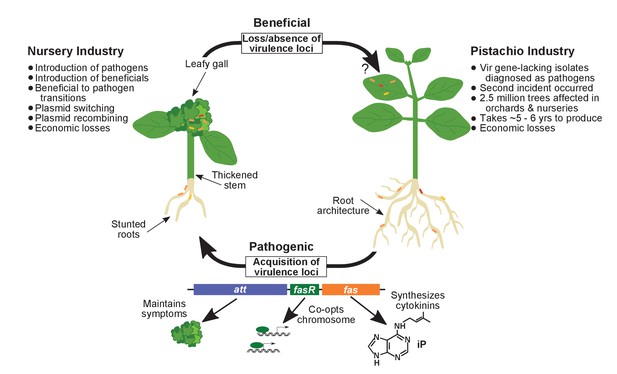
Model of the evolutionary transition in Rhodococcus and effects on agricultural sectors.
The presence or absence of the virulence plasmid in eight species of Rhodococcusdetermines whether the bacterium is beneficial (right), and promotes growth in root architecture, or pathogenic (left), and causes leafy galls and inhibits primary growth. The virulence plasmid carries three loci identified as necessary for pathogenicity and their predicted functions are described. Horizontal gene transfer and evolutionary transitions affect Rhodococcus and impact the nursery industry (as indicated on the left of the figure). Virulence-gene-lacking isolates of Rhodococcus were diagnosed as outbreak strains and the probable misdiagnosis could have had detrimental impacts on the pistachio industry (as indicated on the right of the figure).
Additional files
-
Supplementary file 1
Metadata and other supporting information.
(A)Characteristics of Rhodococcus isolates sequenced in this study. (B) Average nucleotide identity of Rhodococcus. (C) Single nucleotide polymorphisms (SNPs) for Clade I isolates of Rhodococcus. (D) Single nucleotide polymorphisms (SNPs) for Clade II isolates of Rhodococcus. (E) List of 123 genes present in 95% or more plasmid sequences. (F) Characteristics of Rhodococcus isolates used in this study. (G) Genes enriched in Clade I–IV members of Rhodococcus. (H) Polymorphisms between Rhodococcus D188 and D188-5. (I) Sequences of primers and probes used in this study.
- https://doi.org/10.7554/eLife.30925.033
-
Transparent reporting form
- https://doi.org/10.7554/eLife.30925.034





