The P2X7 receptor forms a dye-permeable pore independent of its intracellular domain but dependent on membrane lipid composition
Figures
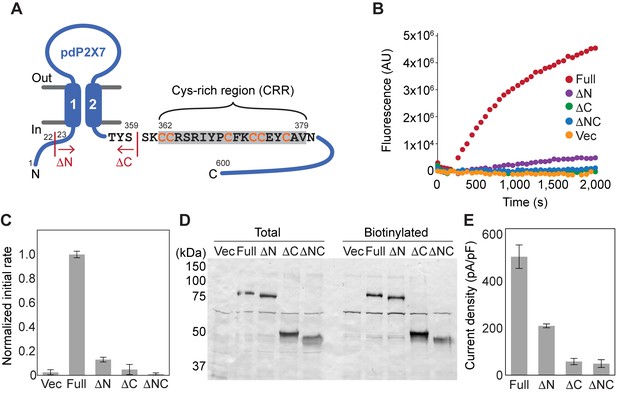
Characterization of deletion constructs of pdP2X7.
(A) Schematic representation of pdP2X7. The start or end position of each deletion construct is shown. ΔN: Δ1–22; ΔC: Δ360–600; ΔNC: Δ1–22/Δ360–600. (B and C) ATP-evoked YO-PRO-1 uptake in HEK293 cells expressing each construct. Representative results are shown in (B). The initial rate of YO-PRO-1 uptake after ATP application was calculated and normalized to the initial rate of the full-length pdP2X7 (C). The bars represent the means of at least five independent experiments and the error bars represent SEM. Vec; vector transfected HEK cells. (D) Surface expression of P2X7 constructs. FLAG tag was attached to the N-terminus of P2X7 constructs and expressed in HEK293 cells. Surface expressed protein was biotinylated and probed by western blotting. Total protein sample after solubilization (total) and eluted sample from resin (biotinylated) are shown. (E) Current densities of the pdP2X7 constructs. Currents were obtained by whole cell patch clamp recordings triggered by 1 mM ATP. Membrane potential was held at −60 mV. Bars represent the means of at least four measurements and the error bars represent SEM.
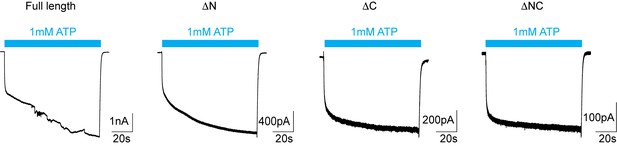
Whole cell patch clamp recordings of pdP2X7 deletion constructs.
Whole cell currents were obtained from HEK293 cells expressing pdP2X7 constructs triggered by 1 mM ATP for 90 s. Membrane potential was held at −60 mV. Note different current scales. Well-clamped recording were unable to be obtained for the full-length pdP2X7, consistent with membrane blebbing triggered by channel activation (Virginio et al., 1999).
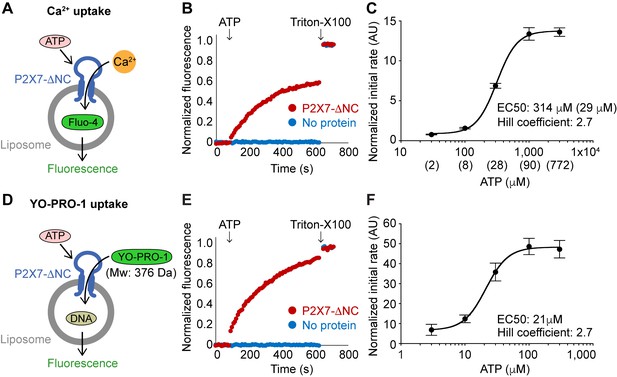
Reconstituted pdP2X7-ΔNC takes up both Ca2+ and YO-PRO-1 in liposomes.
(A) Schematic representation of Ca2+ uptake assay. Purified pdP2X7-ΔNC was reconstituted into liposomes and the Ca2+ indicator Fluo-4 was incorporated by freeze-thaw cycles. Ca2+ influx was monitored after ATP application by measuring Fluo-4 fluorescence. (B) pdP2X7-ΔNC mediated Ca2+ uptake triggered by ATP. pdP2X7-ΔNC was reconstituted into POPE:POPG 3:1 (w/w) liposomes at 1:100 ratio of protein to liposome and Fluo-4 fluorescence was monitored before and after application of 1 mM ATP (red). Fluorescence was normalized to the total fluorescence obtained after application of 1% Triton-X100. Empty liposomes (no protein) with the same composition are shown as a control (blue). (C) ATP dose-responses of pdP2X7-ΔNC in liposome for Ca2+ uptake. Plots were made using the means of six independent experiments and the error bars represent SEM. Dose-response curves were fit to the Hill equation. The numbers in parentheses are calculated free ATP (ATP4−) concentrations using Max Chelator (http://maxchelator. stanford.edu/). (D) Schematic representation of YO-PRO-1 uptake assay. Purified pdP2X7-ΔNC was reconstituted into liposomes and a 20-mer double-strand DNA was incorporated by freeze-thaw cycles. YO-PRO-1 influx was measured by an increase of YO-PRO-1 fluorescence triggered by DNA-binding. (E) pdP2X7-ΔNC mediated YO-PRO-1 uptake triggered by 1 mM ATP. pdP2X7-ΔNC was reconstituted into POPE:POPG 3:1 (w/w) liposomes and YO-PRO-1 fluorescence was monitored before and after application of 1 mM ATP (red). Fluorescence was normalized to the total fluorescence obtained after applying 1% Triton-X100. Empty liposomes (no protein) with the same composition are shown as a control (blue). (F) ATP dose-response of pdP2X7-ΔNC in liposomes determined by YO-PRO-1 uptake. The plots were made using the means of four independent experiments and the error bars represent SEM. Dose response curves were fit to the Hill equation.
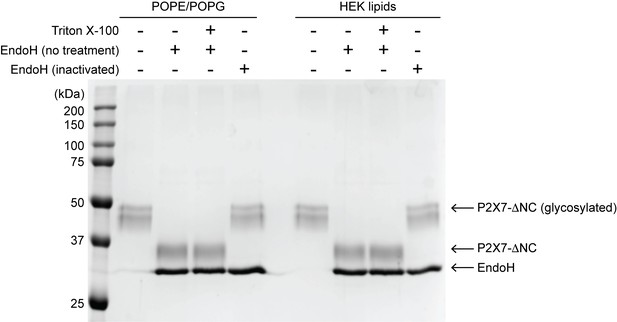
Outside-out configuration of pdP2X7-ΔNC in liposomes.
Proteoliposomes were treated with recombinant EndoH in the presence or absence of Triton-X100. EndoH was inactivated at 75°C for 15 min prior to incubation with SDS-PAGE sample buffer. Pre-heated EndoH did not shift P2X7-ΔNC bands, excluding the possibility that deglycosylation occurred on denatured proteins in SDS-PAGE sample buffer. Proteoliposomes were analyzed on an SDS-PAGE gel stained with Coomassie Blue.
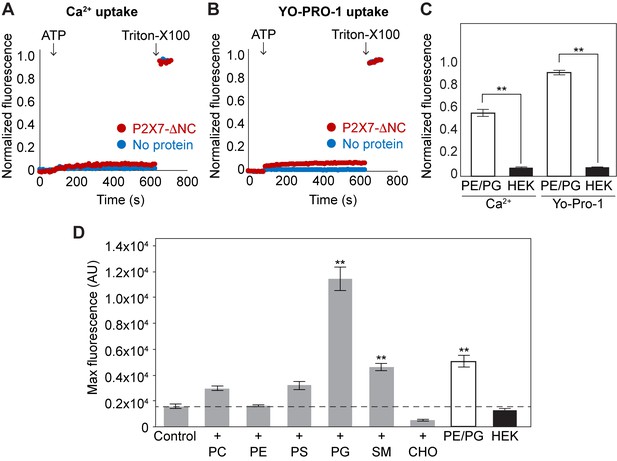
Lipid-dependent YO-PRO-1 uptake of pdP2X7-ΔNC in liposomes.
(A) and (B) Ca2+ and YO-PRO-1 uptake of pdP2X7-ΔNC in HEK293-mimetic liposomes. pdP2X7-ΔNC was reconstituted into liposomes composed of 28.5% POPC, 16.5% POPE, 2.25% POPG,18% POPS, 9.75% sphingomyelin, and 25% cholesterol. Fluo-4 (A) or YO-PRO-1 (B) fluorescence was monitored before and after application of 1 mM ATP (red). Empty liposomes (no protein) with the same composition are shown as a control (blue). Fluorescence was normalized to the total fluorescence obtained after application of 1% Triton-X100. (C) Comparison between Ca2+ and YO-PRO-1 uptake in POPE/POPG or HEK293-mimetic liposomes. The bars represent the means of maximum fluorescence after ATP application normalized to the total fluorescence obtained after application of 1% Triton-X100. The means were obtained from at least four independent experiments and error bars represent SEM. Asterisks indicate a significant difference (p<0.01) between the PE/PG and HEK293-mimetic liposomes as determined by student's t-test. (D) Lipid-dependent YO-PRO-1 uptake of pdP2X7-ΔNC. Purified pdP2X7-ΔNC was reconstituted into liposomes with different lipid compositions. The bars represent the means of maximum fluorescence after ATP application from four independent experiments and the error bars represent SEM. Control: 50% POPE and 50% POPC;+PC: 25% POPE and 75% POPC;+PE: 75% POPE and 25% POPC;+PS: 25% POPE, 25% POPC, and 50% POPS;+PG: 25% POPE, 25% POPC, and 50% POPG;+SM: 25% POPE, 25% POPC, and 50% sphingomyelin;+CHO: 25% POPE, 25% POPC, and 50% cholesterol; PE/PG: 75% POPE and 25% POPG; HEK: 28.5% POPC, 16.5% POPE, 2.25% POPG,18% POPS, 9.75% sphingomyelin, and 25% cholesterol. Asterisks indicate significant difference compared to the control (p<0.01) as determined by one way ANOVA followed by Dunnett's test.
Comparable reconstitution efficiency of pdP2X7-ΔNC in liposomes with different lipid compositions.
SDS-PAGE image of proteoliposomes used in Figure 3D.
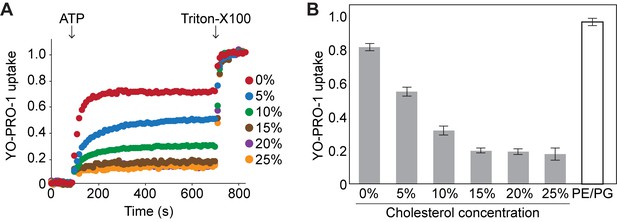
Cholesterol inhibits YO-PRO-1 uptake of pdP2X7-ΔNC in a dose dependent manner.
(A) Purified pdP2X7-ΔNC was reconstituted into liposomes with different cholesterol compositions and ATP-evoked YO-PRO-1 uptake was measured. Liposomes were prepared by adding different amounts of cholesterol (final 5–25%) to the lipid mixture composed of 38% POPC, 22% POPE, 3% POPG, 24% POPS, and 13% sphingomyelin. (B) Comparison of YO-PRO-1 uptake of proteoliposomes composed of different cholesterol contents. Bars represent the mean maximum fluorescence after ATP application from nine independent experiments and the error bars represent SEM. Fluorescence was normalized to the total fluorescence obtained after application of 1% Triton-X100. PE/PG: liposome containing 75% POPE and 25% POPG.
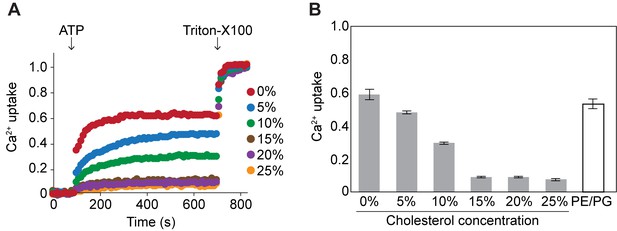
Cholesterol also inhibits Ca2+uptake of pdP2X7-ΔNC in a dose dependent manner.
(A) ATP-evoked Ca2+ uptake was measured using proteoliposomes as described in Figure 4. (B) Bars represent the mean maximal fluorescence after ATP application from four independent experiments and the error bars represent SEM. Fluorescence was normalized to the total fluorescence obtained after application of 1% Triton-X100. PE/PG: liposome containing 75% POPE and 25% POPG.
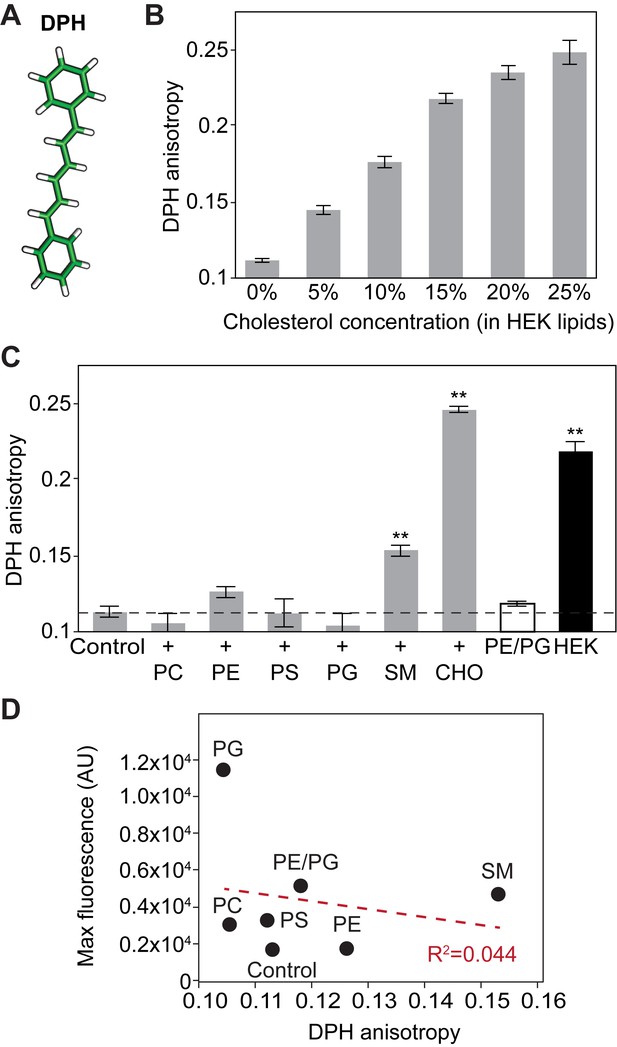
Membrane rigidity of liposomes composed of different lipids monitored by DPH fluorescence anisotropy.
(A) Stick representation of 1,6-diphenyl-1,3,5-hexatriene (DPH). (B) DPH anisotropy from pdP2X7-ΔNC reconstituted liposomes composed of HEK lipids with designated amount of cholesterol. DPH anisotropy was obtained at Ex: 358 nm and Em: 429 nm. (C) DPH anisotropy from pdP2X7-ΔNC reconstituted liposomes composed of different kinds of lipids as described in Figure 3. Asterisks indicate significant difference compared to wildtype or the no protein control (p<0.01) as determined by one-way ANOVA followed by Dunnett's test. (D) Linear regression analysis of maximum fluorescence and DPH anisotropy. The shown values are the averages of 4–5 independent experiments.
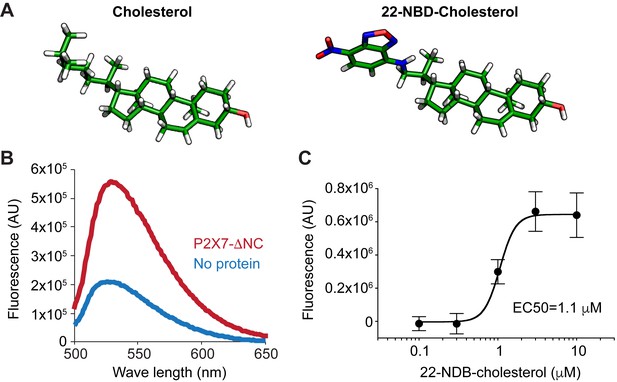
Cholesterol binds to purified pdP2X7-ΔNC.
(A) Stick representation of cholesterol and 22-NBD-cholesterol. (B) Fluorescence spectra of 22-NBD-cholesterol after incubating with pdP2X7-ΔNC (red) or without protein (blue) for 1 hr at room temperature. (C) Dose-dependent 22-NBD-cholesterol binding to pdP2X7-ΔNC. The plots were made using the means of five independent experiments and the error bars represent SEM. Dose response curves were fit with the Hill equation.
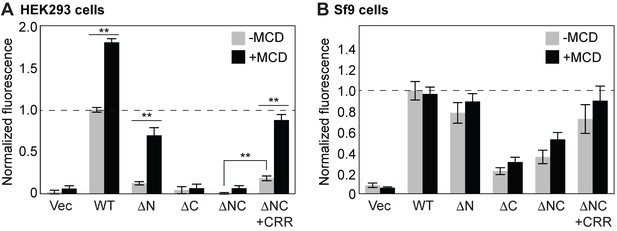
CRR facilitates YO-PRO-1 uptake in the presence of MCD.
(A) ATP-evoked YO-PRO-1 uptake from HEK293 cells expressing each construct. HEK293 cells were treated with 5 mM Methyl-β-cyclodextran (MCD) for 1 hr at 37°C before the assay was performed. The initial rate of YO-PRO-1 uptake in the presence (black) or absence (grey) of MCD was calculated and normalized by the initial rate of the full length protein without MCD. The bars represent the means of at least five independent experiments and the error bars represent SEM. (B) ATP-evoked YO-PRO-1 uptake from Sf9 cells expressing each construct. Bars represent the means of the normalized initial rate from five independent experiments and the error bars represent SEM. Asterisks indicate significant difference between YO-PRO-1 uptake with and without MCD treatment (p<0.01) as determined by one way ANOVA followed by Dunnett's test. No significant fluorescence change was observed for YO-PRO-1 uptake from Sf9 cells with or without MCD.
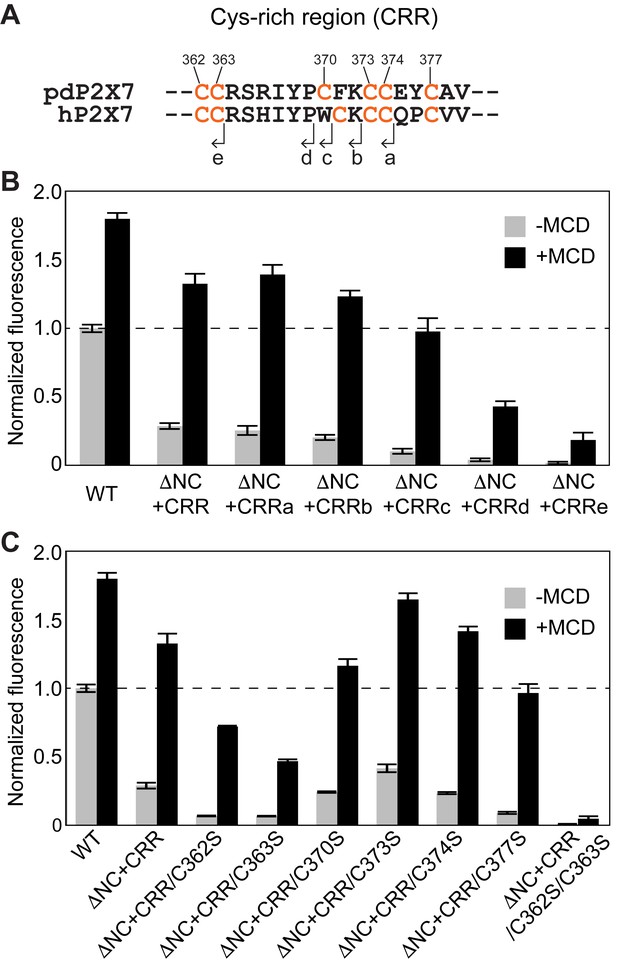
Critical region of CRR for facilitating P2X7 mediated YO-PRO-1 uptake.
(A) Amino acid sequences of the CRR of panda and human P2X7 receptors. The end position of each deletion construct (CRRa-e) is shown. (B) ATP-evoked YO-PRO-1 uptake from HEK293 cells expressing each pdP2X7 deletion construct in the presence (black) or absence (grey) of MCD. Initial rate of YO-PRO-1 uptake was calculated and normalized by the initial rate of the full length protein without MCD. The bars represent the means of at least eight independent experiments and error bars represent SEM. (C) ATP-evoked YO-PRO-1 uptake of HEK293 cells expressing Cys knockout mutants in the presence (black) or absence (grey) of MCD. The bars represent the means of at least four independent experiments and the error bars represent SEM.
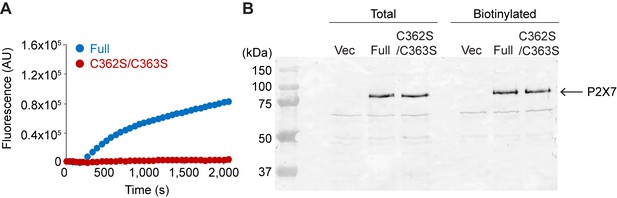
C362S/C363S mutation abolishes YO-PRO-1 uptake of pdP2X7 in HEK293 cells.
(A) YO-PRO-1 uptake of full length and the C362S/C363S mutant. Representative trace of eight independent experiments is shown. (B) Surface expression of the wildtype and the C362S/C363S pdP2X7 mutant. FLAG tag was attached to the N-terminus and expressed in HEK293 cells. Surface expressed protein was biotinylated and cells were lysed. Biotinylated protein was collected by streptactin resin. Total protein sample after solubilization (total) and eluted sample from resin (biotinylated) were applied to SDS-PAGE and immunoblotting was performed using FLAG antibody.
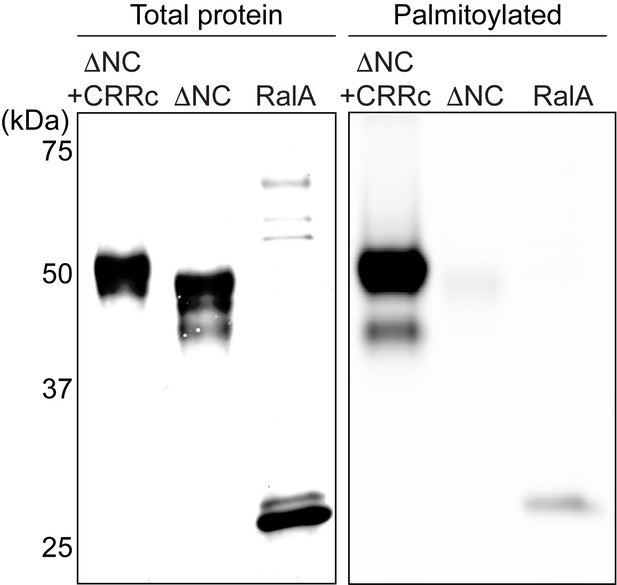
CRR of pdP2X7 is palmitoylated in HEK293 cells.
HEK293 cells transfected with FLAG-pdP2X7-ΔNC+CRRc, FLAG-pdP2X7-ΔNC, or FLAG-RalA were metabolically labeled with 17-ODYA and pulled down using anti-FLAG antibody. The amount of loaded protein (total protein) was detected by western blotting using anti-FLAG antibody and 17-ODYA incorporation was detected with Alexa Fluor 647-azide (Palmitoylated).
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31186.016