Pi-Pi contacts are an overlooked protein feature relevant to phase separation
Figures
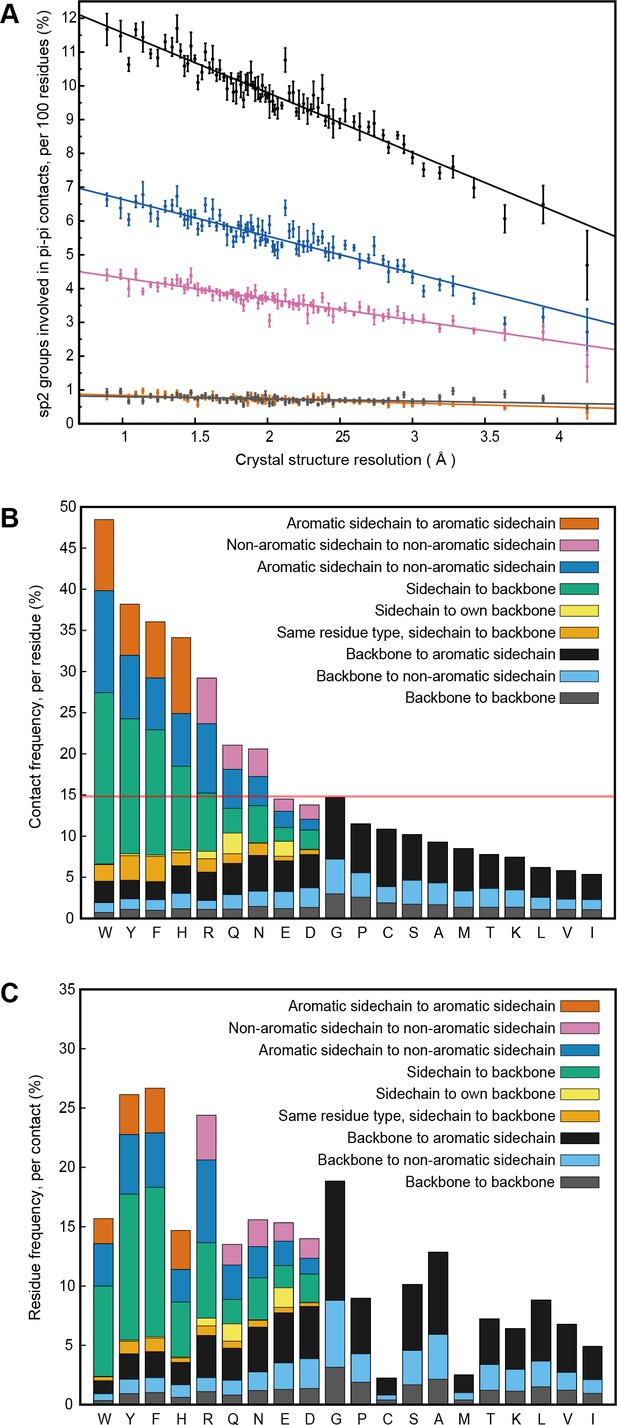
PDB statistics for planar pi-pi interactions.
(A) Average number of sp2 groups involved in planar pi-pi contacts per 100 protein residues binned by crystal structure resolution. Values are shown for contacts defined by the nature of the involved sp2 groups, with all groups in black, aromatic to non-aromatic sp2 in blue, non-aromatic to non-aromatic in pink, backbone to backbone in gray, and aromatic to aromatic in orange. Error bars show bootstrap SEM. (B) Planar pi-pi contact interaction frequencies for each residue type, with the average across all residue types shown as a red line, and (C) frequency of each residue type in contributing to planar pi-pi interactions, with bars showing overall frequency colored proportionally by the nature of the contact partners. Figure 1—source data 1 and 2.
-
Figure 1—source data 1
Pi-Pi contact annotations for the full PDB set.
Text file listing the pi-pi contacts observed across our non-redundant PDB set, with contact types shown by residue annotations where single amino acid names refer to sidechains and pairs of amino acids refer to the backbone peptide bond between residue i and residue i + 1.
- https://doi.org/10.7554/eLife.31486.005
-
Figure 1—source data 2
Residue and amino acid counts for the full PDB set.
Text file listing the residues assessed in each individual PDB chain, used for calculating contact frequencies.
- https://doi.org/10.7554/eLife.31486.006
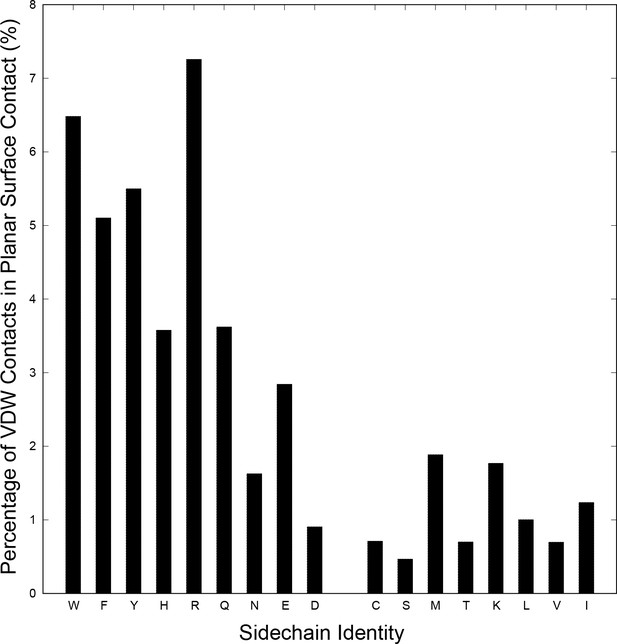
Proportion of sidechain to backbone VDW contacts that satisfy planar contact criterion.
To examine relative contact enrichment, sidechain contacts to the backbone are normalized against the total number of contacts satisfying the same VDW criterion (two pairs of atoms within 4.9 Å), with comparison between (left) planar sp2 sidechain groups (for W, F, Y, H, R, Q, N, E and D) and (right) selected sp3 planar surfaces (for C, S, M, T, K, L, V, I). The sp3 planar surfaces were chosen as a control by taking sets of atoms describing exposed planar surfaces, as described in the Materials and methods. Comparing relative planar contact frequency, we observe the majority of sp2 sidechain types show clear enrichment relative to the sp3 controls.
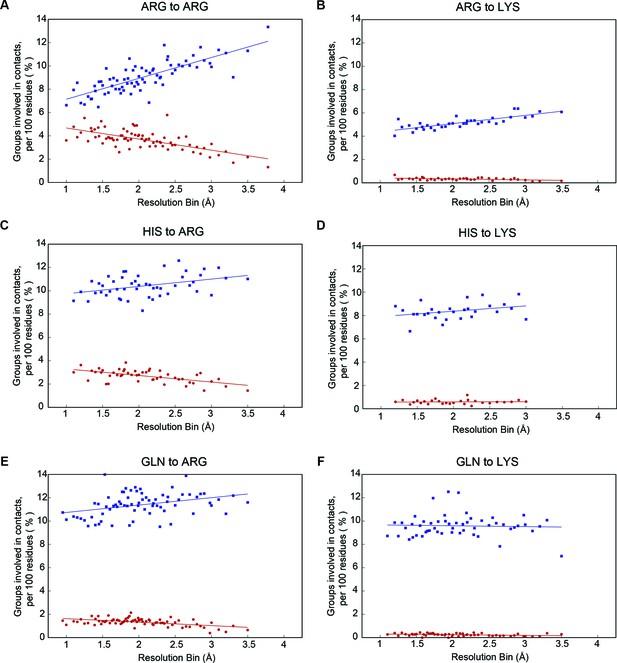
Selected sidechain-to-sidechain contact frequencies by resolution.
Percentage of residues involved in planar contacts are shown in red, and percentage in any other non-planar VDW contact are shown in blue, with panels showing contacts by sidechain group (for panels A-F: R to R, R to K, H to R, H to K, Q to R, and Q to K). We observe that the increase in planar pi-pi contacts to arginine at higher resolution comes at the expense of non-planar VDW contacts (panels A, (C and E). In contrast, contacts made to an arbitrary surface plane at the end of lysine sidechains do not show this increase in planar orientation with resolution (panels B, D and F).
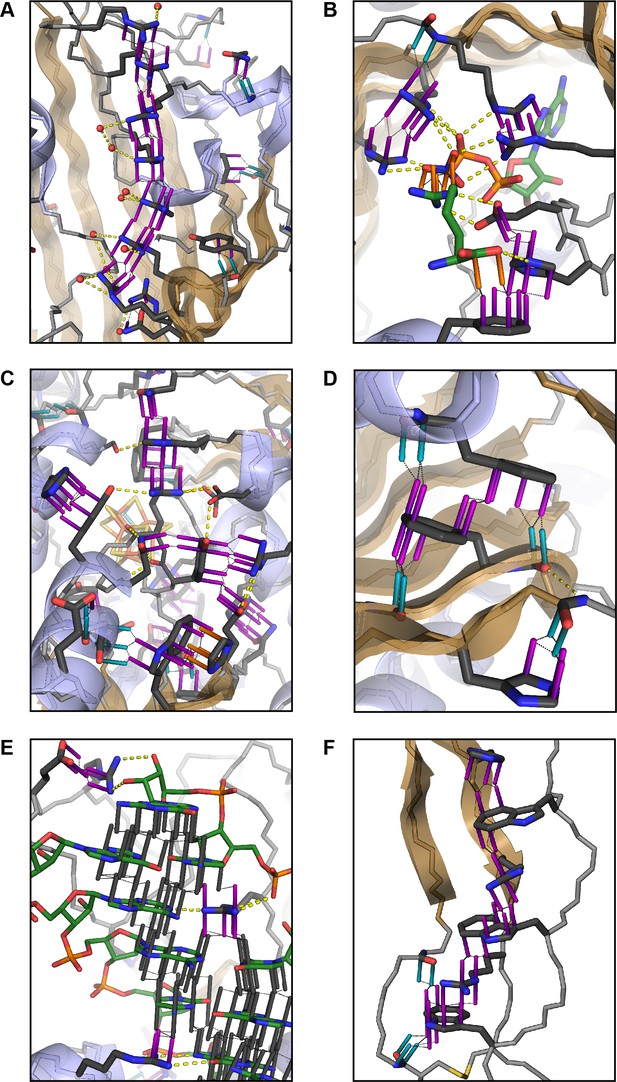
Examples of planar pi-pi contacts in folded protein structures.
Pi-pi interactions shown using rods to describe the normal vector of the plane. Rods extend to a carbon VDW radius of 1.7 Å, colored by category with sidechain groups in purple, backbone in blue, small molecule ligands in orange, and RNA in gray. Ligand molecules are green, with relevant water molecules shown as red spheres and hydrogen bonds as yellow lines. (A) Arginine ladder motif in Porin P (PDB:2o4v). (B) Catalytic site from arginine kinase (PDB:1m15). (C) Network of interactions in nitrogenase (PDB: 3u7q). (D) Backbone/sidechain contacts at the ends of secondary structure elements (PDB:4b93). (E) RNA-binding interactions (PDB: 4lgt). (F) Interaction network stacked between disulfide bonds (PDB: 4v2a).
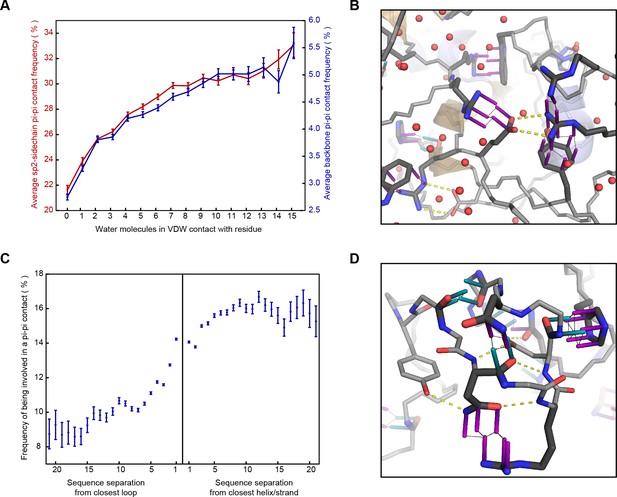
Correlation of planar pi-pi interactions with solvent and lack of secondary structure.
(A) Contact frequency for sidechain groups (red) and backbone (blue) increases with the total number of solved water molecules within 4.9 Å of the residue, based on structures with >1 water oxygen per residue, including all molecules within 8 Å of the chain of interest, including symmetry partners. (B) Representative example of a pi-stacked sidechain in contact with 11 water molecules (PDB:4u98), showing how the interaction does not appear to compete with solvent. (C) Mean contact frequency vs. sequence distance from regular secondary structure and loop/turn regions. (D) Example of the range of interactions found >10 residues from helix/strand secondary structure (PDB:4b4h).
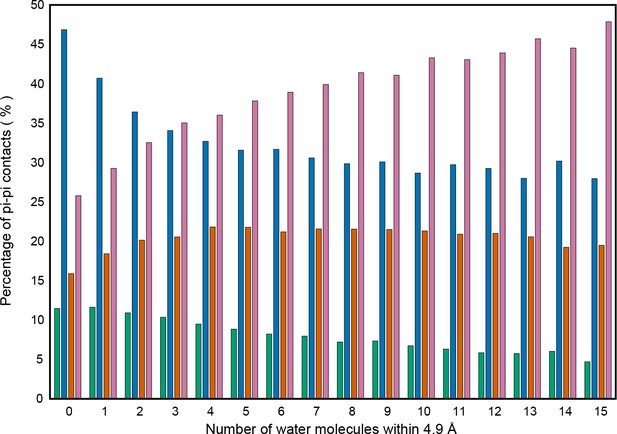
Effect of solvation on pi-pi category frequencies.
Effects of solvation, measured by the total number of water molecules within 4.9 Å of a given residue, on the overall frequency of different types of interactions, categorizing contacts by the identities of the solvent contact tested residue and its partner, where the solvated residue is listed first (green for aromatic to aromatic, blue for aromatic to non-aromatic, orange for non-aromatic to aromatic, and pink for non-aromatic to non-aromatic). Note that non-aromatic includes backbone interactions.
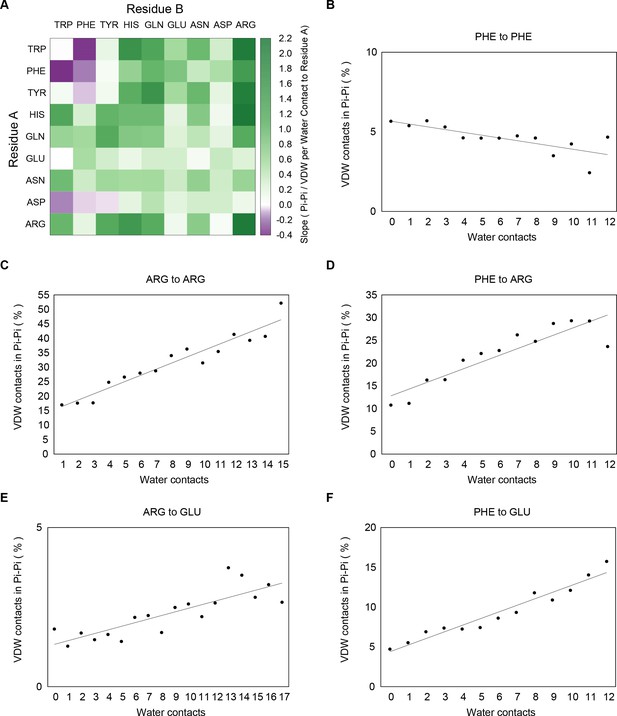
Enrichment of pi-pi contacts, relative to overall VDW contacts, as a function of the number of interactions with water.
Water contacts are measured to residue A, and the percentage of pi-pi contacts per VDW contact is measured for all contacts from residue A to residue B. Panel A shows the change in percentage of pi-pi contacts per VDW contact by number of waters for each sidechain-sidechain interaction, with pi-contact enrichment with solvation being a consistent property of the majority of interactions involving at least one non-aromatic sidechain. Panels B-F show slope measurements for a selection of examples, Phe to Phe, Arg to Arg, Phe to Arg, Arg to Glu and Phe to Glu, respectively.
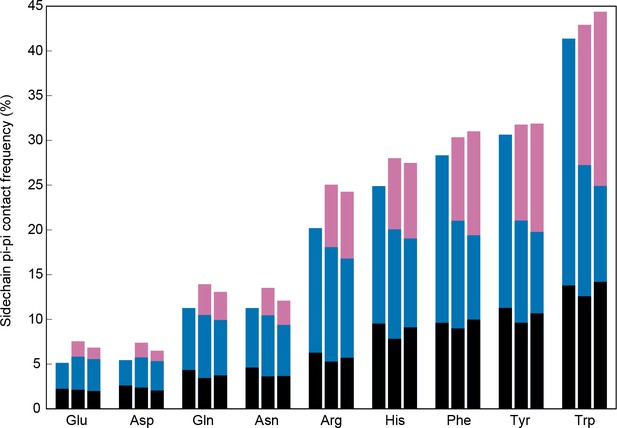
Sidechain contacts at interface positions.
Contact frequencies are shown for the nine sp2-containing sidechain types, split into three bars based on interface proximity. From left to right, these bars are i) no other chain within 4.9 Å of any sidechain atom, ii) within 4.9 Å VDW contact distance of any atoms in a different chain within the unit cell of the crystal, iii) within 4.9 Å of any atoms in a chain from a neighboring unit cell, as determined by crystal symmetry data. Bars are colored by the proportion of total contacts contributed by three categories, bottom/black corresponding to local (sequence separation ≤4 residues) intrachain contacts, middle/blue to non-local intrachain contacts, and top/pink to interchain contacts, showing that overall contact frequencies and local contact frequencies remain similar and that the non-local contacts do not discriminate between intra and interchain.
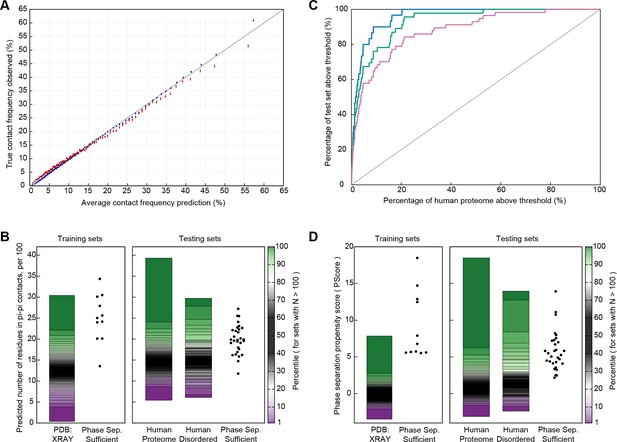
Prediction of phase separation based on planar pi-pi interactions.
(A) Reliability plot showing average predicted and observed contact frequencies for percentile bins by pi-pi contact prediction for proteins in the PDB, with PDB sequences used for training in blue and the leave out set in red. Bars show SEM. (B) Highest number of contacts predicted, by window, for two phase separation predictor training sets and three test sets, for the unoptimized predictor. (C) Modified ROC curve showing the final predictor’s performance on three test sets vs. the human proteome, with the full set in pink (N = 62), the full set minus the insufficient for phase separation set shown in green (N = 44), and the sufficient for phase separation set in blue (N = 32). (D) Results for the final predictor (as for panel b) plotted with the predictor’s phase separation propensity scores (PScore). Data underlying B-D included in Figure 5—source data 1 and Figure 5—source data 2.
-
Figure 5—source data 1
Phase separation training, testing and designed protein test sets.
Excel table containing identification and literature references for proteins in the phase separation test and training sets, with sheet one showing the training set proteins, two showing proteomic test set proteins, and three showing synthetic test set proteins.
- https://doi.org/10.7554/eLife.31486.015
-
Figure 5—source data 2
Additional phase separation propensity scores used in final ROC analysis.
Excel table containing protein IDs and predicted propensity scores, with different datasets on each sheet. Sheets 1–3 have full predictions for the human, E. coli, S. cerevisiae proteomes, respectively. Sheet four repeats the subset of human proteins found in the DisProt database. Sheet five shows scores for the protein sequences found in our non-redundant PDB set, and sheet six repeats the subset of PDB sequences withheld from predictor training.
- https://doi.org/10.7554/eLife.31486.016
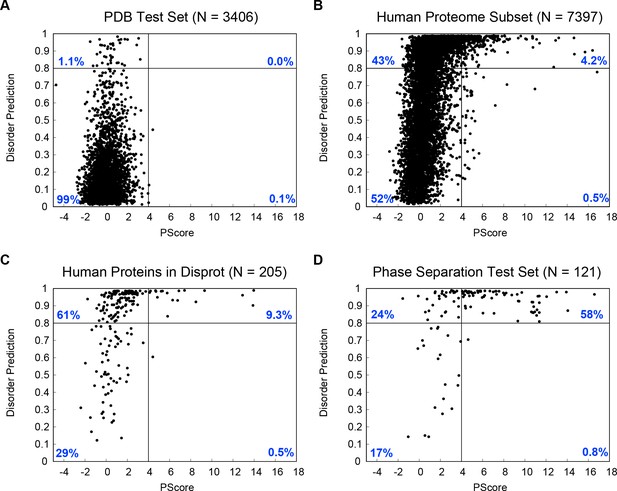
Contrasting behavior of disorder prediction algorithms and the phase separation prediction.
Disopred3 (Jones and Cozzetto, 2015) derived disorder predictions are shown on the y axis and PScores are shown on the x axis for four different test sets, (A) our PDB test set, representing a negative set for both phase separation and disorder, (B) a random sample of 4385 sequences from the human proteome, (C) the subset of the human proteome annotated as containing disorder in the Disprot database (Piovesan et al., 2017), representing a positive set for disorder, and (D) our full phase separation test set. Results are split into four categories separated by PScore = 4 and Disorder = 0.8, with the percentage of sequences in each category inset in blue. The majority of known phase-separating proteins are associated with disorder, and are predicted to be disordered, but sequences predicted to phase separate represent a small subset of both the known and the predicted disordered proteins.
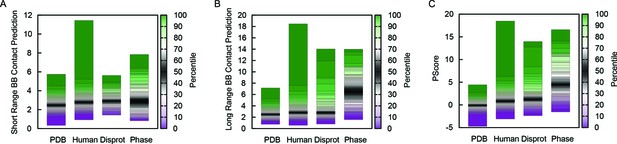
Comparison of scores used in generating phase separation predictions.
(A) Highest number of short-range backbone contacts predicted, by window, for the PDB test set, the human proteome, the set of disordered human proteins from Disprot, and the full phase separation test set (N = 121), where percentile ranges are shown in colored boxes. (B) Highest number of long-range backbone contacts predicted, as for panel a. (C) Results for the final predictor plotted with the predictor’s phase separation propensity scores (PScore). Prediction of long-range backbone contacts provides the majority of the discrimination seen in the final predictor.
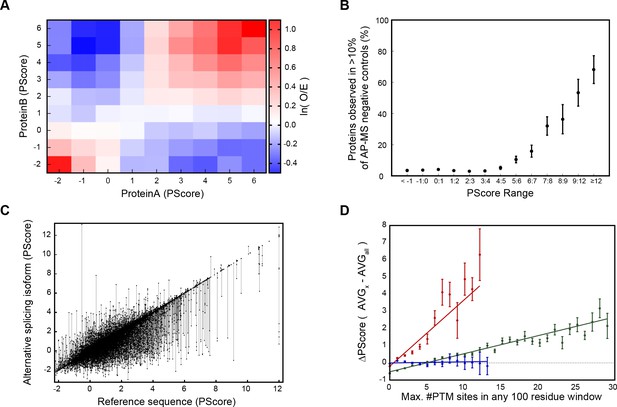
Association of phase separation propensity scores with protein interactions, splice isoforms, PTMs, and GO localization, process, and function terms.
(A) Protein-protein interaction enrichment by the PScore of partner 1 vs. the PScore of partner 2. The color gradient shows the natural logarithm of the observed over expected ratio. (B) Percentage of human proteins at each PScore range that are detected in more than 10% of AP-MS negative control experiments. (C), Score ranges for alternative splicing variants shown as vertical lines sorted by reference sequence values. (D), Number of PTMs vs. average relative PScore, with methylation shown in red, phosphorylation in green, and ubiquitination in blue.
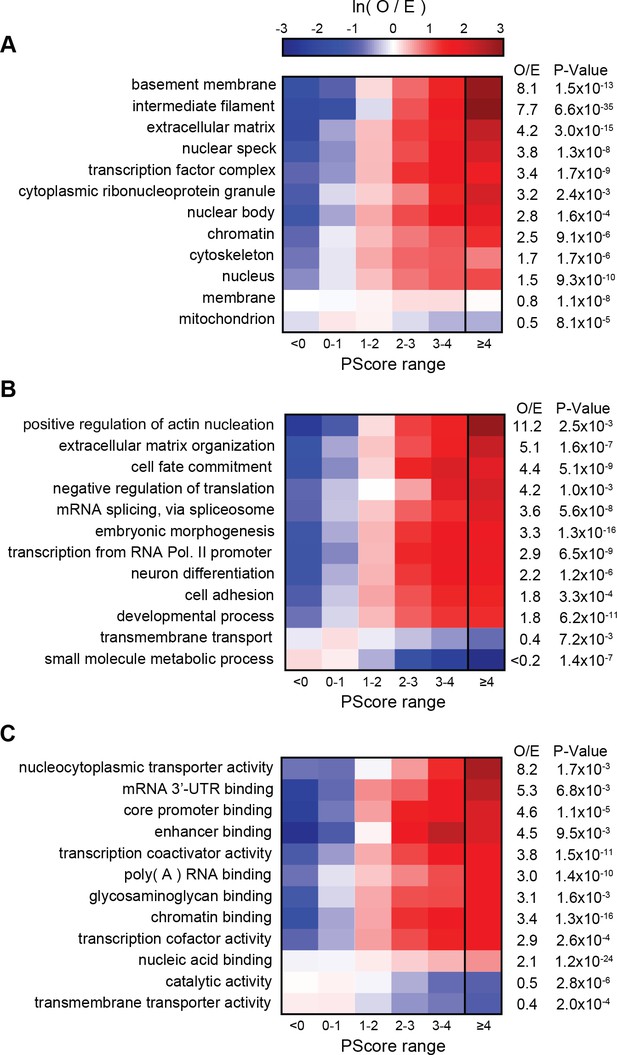
PScore enrichment by gene ontology annotation for subcellular localization (A), biological process (B), and molecular function (C).
The color gradient shows the natural logarithm of the observed over expected ratio. Heatmaps show enrichment in vertebrate sequences across six defined score ranges, with the highest score range (PScore ≥4) labeled with human enrichment values calculated using PANTHER (see Materials and methods).
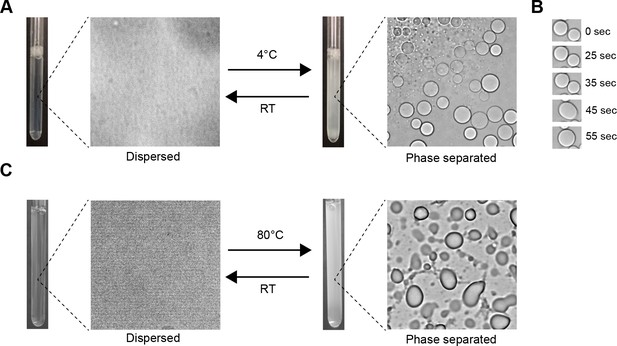
Visual confirmation of phase separation.
(A) Test tubes containing transparent or turbid solutions of 1 mM FMR1 C-terminus (residues 445–632) along with their corresponding DIC microscopy images taken at room temperature or 4°C, respectively. (B) 1 mM FMR1 C-terminus forms droplets exhibiting liquid fusion properties at 4°C. (C) 40 µM solutions of Human Cytalomegalovirus pAP along with corresponding microscopy images taken at room temperature or 80°C, respectively.
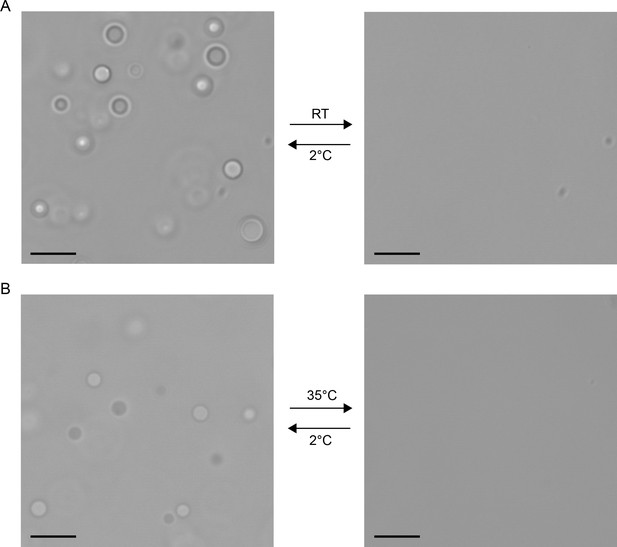
Visual confirmation of phase separation, using 20 mg/ml ficol as a crowding agent.
(A) 200 µM FMR1 C-terminus shows reversible droplet formation between 2°C and RT, (B) 220 µM engrailed-2 shows reversible droplet formation between 2°C and 35°C. DIC Images taken at 63x magnification, where shading reflects the differences in position relative to the focal plane of the free floating droplets. Scale shown as black bars sized to 10 µm.
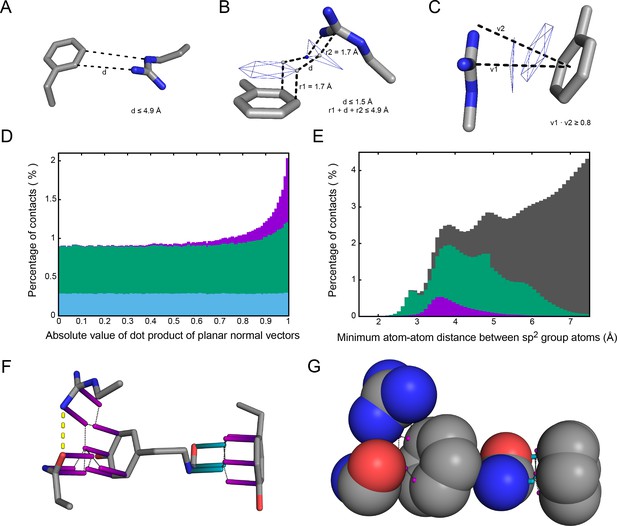
Contact definitions.
(A) Contacts are identified first as sp2 planes in which at least two pairs of atoms come within 4.9 Å of one another, and then by restricting to the subset with (B) planar surfaces (at the carbon VDW radius of 1.7 Å) with points along the planar normal vectors coming within 1.5 Å of one another and (C) a planar orientations for which the absolute value of the dot product of normal vectors is ≥0.8. (D) Shows the rationale for these restrictions, where binning sidechain-sidechain interactions by the relative orientation between planes shows that planar (same-orientation) interactions, primarily in the 0.8 to 1.0 range (angles between the planes from 0 deg to 36 deg), show enrichment relative to the uniform distribution expected for random orientations. Of these, interactions with only one atom-atom pair within VDW contact (shown in blue) have no bias. Enrichment comes entirely from contacts with either two pairs of planar surfaces within 1.5 Å of each other (shown in purple) or two distinct pairs of atoms within 4.9 Å but without the planar surface contact (shown in green). (E) Minimum distance measurements between pairs of atoms found in separate sp2 groups, measured from the closest pairing for each atom. Gray shows all sidechain-sidechain measurements, and green/purple show distances corresponding to the groups in D. (F) Representative examples of sidechain-sidechain and sidechain-backbone pi-contacts are shown as sticks (PDB: 1gde), with carbon atoms in gray, oxygen in red, and nitrogen in blue. Planar normal vectors extended to the carbon VDW radius, representing pi-orbital locations, are shown as purple rods for sidechain groups and blue rods for backbone groups, and the yellow line denotes a hydrogen bond where both donor and acceptor atoms are in pi-contact distance to a third sidechain. (G) A space-filling representation of the sp2 atoms in F, with gray lines between normal vector rods used to show the planar surface measurements taken for defining pi-contacts.
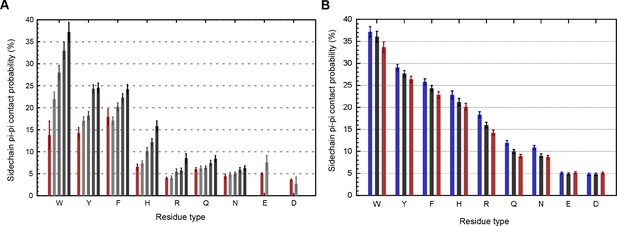
Cross validation against NMR restraints and X-ray structure resolution.
(A), The relationship between contact frequency and experimental data quality is not unique to crystallography, as shown by the effect of increasing the number of restraints on sidechain specific contact frequencies over 2589 structures solved by NMR. For each sidechain/protein combination we calculated the average number of distance restraints involving sidechain atoms (from the first sp2 atom onward), and then binned residues into five categories, with red for structures without any sidechain distance restraints for that residue type, and ranking quartiles from light gray to black by order of increasing restraints, where the consistent increase in contact frequency from left to right confirms that more restraints result in higher planar pi-contact frequencies. For Glu and Asp, less than 1% of the structures were derived using distance restraints to the carboxyl's lone carbon atom so we did not split them into quartiles. (B), To control for potential sample bias we also tested the relationship between resolution and contact frequency for crystallographic structures that have been solved at least three different times at different resolutions, with bars showing contact frequencies over identical populations of residues for the highest (blue), median (black), and lowest resolution (red) structures. Error bars show standard error of the mean (SEM).
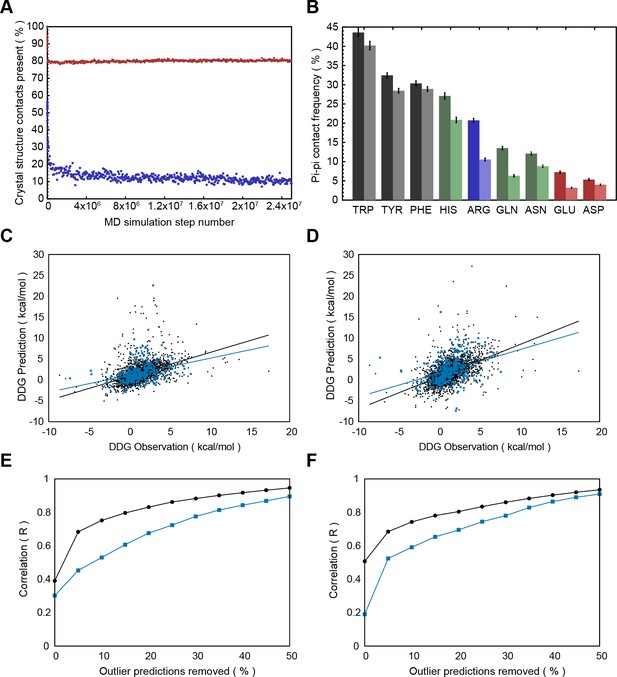
Pi-pi interactions underestimated by some energy functions.
(A), Contact frequency during molecular dynamics simulations of 100 proteins, made available through Dynameomics (Kehl et al., 2008), shows a rapid initial loss of >80% of sidechain pi-contacts which continues to decline throughout the simulation (blue points). By comparison, sidechain hydrogen bonding shows a stable loss of only 20% of interactions (red points). (B), Minimization of 762 crystal structures against the Talaris2014 energy function by Rosetta3.4 (Leaver-Fay et al., 2011; O'Meara et al., 2015), with starting contact frequencies (left bars) decreasing after minimization (right bars). (C–F), Analysis of the relationship between the energetic effects of point mutations (ΔΔG) and pi-contacts for experimental ΔΔGs (blue bars) and ΔΔGs predicted by simulation against the FOLDX force field (Schymkowitz et al., 2005) (C,E) and Rosetta (D,F). Panels C,D show predicted ΔΔG values vs. observation for residues that are not involved in pi-contacts in black, and residues that are involved in pi-contacts in blue, with lines of best fit colored the same. Panels E,F show how correlation values change as outliers are removed, with correlation consistently worse for mutations involving pi-contacts (blue lines) relative to those that don’t (black lines).
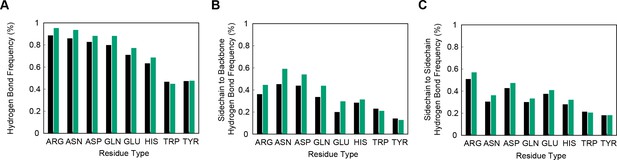
Hydrogen bonding correlates with planar-pi contacts.
Percentage of sidechains involved in at least one hydrogen bond is shown for sidechains that are not in a planar-pi contact in black, and for sidechains that are in a planar-pi contact in green, with panel (A) showing the hydrogen bond frequency across all groups, including ligands and water, (B) showing the hydrogen bond frequency to backbone atoms, and (C) showing the frequency of hydrogen bonding to a sidechain. Hydrogen bond frequency consistently increases with planar pi-pi contacts for all sidechains but Trp and Tyr.
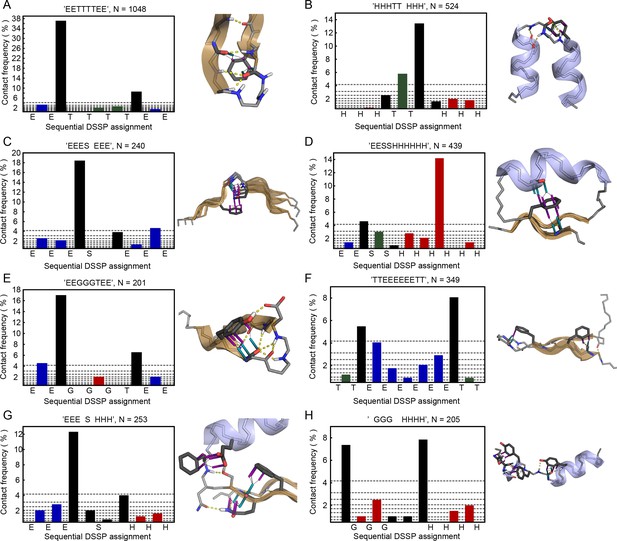
Backbone pi-pi contacts in secondary structure motifs.
Examples of secondary structure motifs showing enrichment for local backbone pi-contacts (contacts made to sidechains within 5 residues of the peptide bond) are displayed. Bar graphs show contact frequency at each position in a motif, as defined by DSSP (Kabsch and Sander, 1983) abbreviated residue class ('E', 'S', 'T', 'H', 'G', and ' '), with bars colored by the associated residues, with green for peptide bonds between two residues classified as turns, blue for bonds in strands, red in helices, and black for bonds that are either unclassified or present at the transition point between classifications. Gray horizontal lines represent the decile values across all backbone contact frequencies, showing that the bonds most likely to end up in the top decile come primarily from transition points between secondary structures (ranging from 2x to 20x enrichment, relative to the median of 1.7%). Protein structures show representative examples of each motif with contacts found at the most enriched position, taken from (A), PDB:1aap, (B), PDB:1gte, (C), PDB:1k5c, (D), PDB:1nhc, (E), PDB:1k3i, (F), PDB:1i8k, (G), PDB:2c4w, and (H), PDB:1kwf.
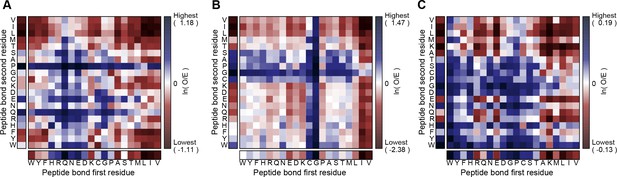
Peptide sequence effects on contact frequency.
Heatmaps show enrichment in the total proportion of planar pi-pi contact involvements observed for peptide bonds between two residues (the first, N-terminal residue on the x-axis and the second, C-terminal residue on the y-axis) relative to the proportion of peptide bonds. Enrichment for (A) short-range contacts (sequence separation <5) and (B) long-range contacts (separation ≥5 or a different chain), respectively, to the peptide bond itself. (C), Enrichment for finding residues within 5 residues of a sidechain that makes a pi-contact to any group in the structure, demonstrating general sequence effects on the contact propensity of neighboring residues. The color gradient shows the natural logarithm of the observed over expected ratio.
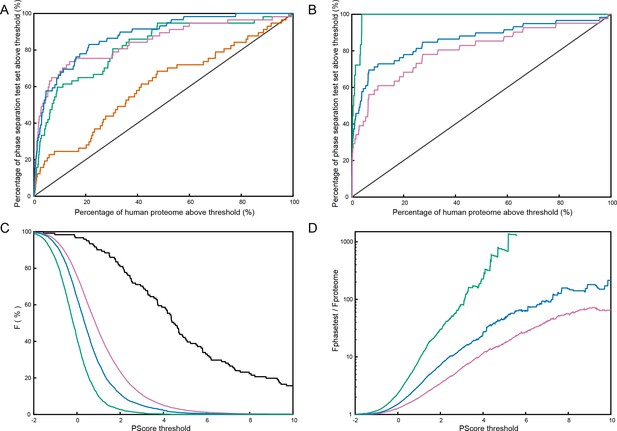
Phase separation propensity predictor testing.
(A), ROC curve comparisons of predictor quality for scores made at different points during the training process, measuring ranking against the full test set (N = 62) vs. the human proteome (only sequences with length ≥140) with green showing the results for the highest number of pi-contacts predicted for any 100 residue window, without any weighting for type (AUC:0.82 ± 0.03), pink and orange showing the same measurement split between long-range (AUC:0.85 ± 0.03) and short-range contacts (AUC:0.62 ± 0.04), respectively, and blue showing the final predictor, which uses weighted combinations of both short- and long-range contact predictions (AUC: 0.88 ± 0.02). (B), the final score tested against 59 phase-separating sequences designed by the Chilkoti lab (Quiroz and Chilkoti, 2015; MacEwan et al., 2017; Simon et al., 2017) (detailed in Figure 5—source data 1C), with comparisons against the full set shown (N = 59) in blue (AUC: 0.86 ± 0.03), and then split into green for 18 proteins shown to phase separate from soluble to insoluble as temperature decreases (AUC:0.99 ± 0.01) and pink for the remaining 41 proteins which phase separate from soluble to insoluble as temperature increases (AUC:0.80 ± 0.04). (C), Fraction of sequences at or above a given PScore, with the combined pool of phase separation test set proteins (N = 121), in black, being compared to three reference proteome sets, with human in pink, S. cerevisiae in blue, and E. coli in green. (D), Enrichment plot for data shown in (C), with ≥PScore frequency for the test set shown relative to proteome frequencies. Analysis based on Figure 5-source data 1 and 2.
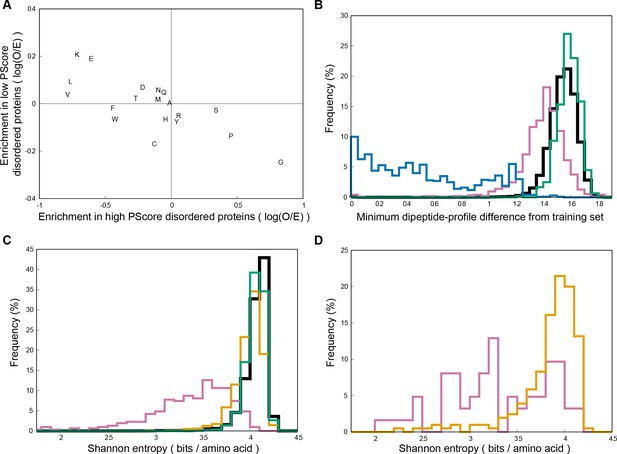
Sequence comparisons of high PScore proteins.
Panel (A) shows compositional bias, relative to the human average, for the high PScore disordered proteins (x-axis) and low PScore disordered proteins (y-axis) used in panel B. High PScore disordered proteins are enriched primarily in Pro and Gly, while low PScore disordered proteins are not enriched in either, but enriched primarily in Lys and Glu, matching our observation that Arg to Lys mutations abrogate phase separation propensity. Panel (B) shows similarity to the training set measured by minimum dipeptide profile distance to any training set protein, as described in the methods. High PScore (≥4.0) human sequences (in pink) are on average closer to the training set than are all human proteins (in black) or PDB sequences (in green), but the range overlaps with both, and is distinct from the similarity seen in blast level homologs of the training set (in blue). Panel (C) shows Shannon entropy distributions of the human proteome (in black), the PDB (in green), and of a set of human proteins proteins predicted to have long stretches of disorder (Disprot3 ≥0.8) split into those with high PScore (≥4, N = 310) (in pink) and low PScore (<1.0, N = 1044) (in orange), showing that PScore but not disorder results in a bias towards lower sequence entropy, suggesting a compositional bias in phase-separating sequences. Panel (D) shows Shannon entropy values for our natural-protein phase separation test set (N = 62) in pink and the disorder-containing human proteins found in Disprot (N = 205) in orange, confirming the observation in panel C that lower Shannon entropy sequences are associated with phase separation.
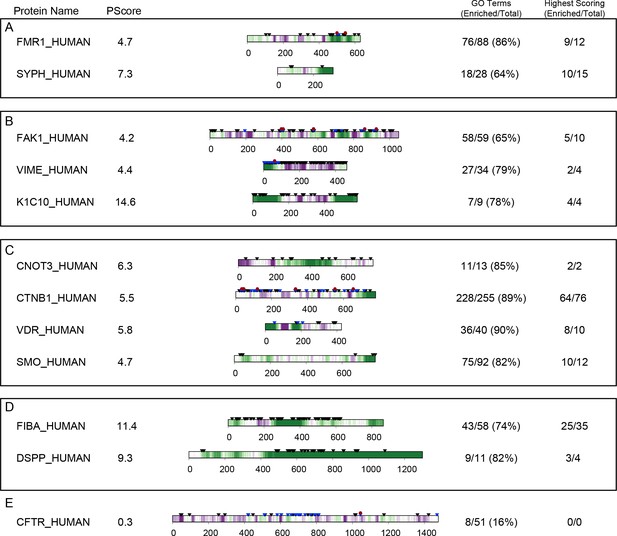
Prediction examples.
Per-residue PScores used to calculate the final full sequence PScore are shown for a selection of human proteins, with residues colored from purple (PScore ≤ −2) to white (PScore = 0) to green (PScore ≥4.0). Black triangles denote residues annotated by PhosphoSitePlus as targets of PTMs, blue triangles denote modification sites with known regulatory significance, and red circles denote modification sites with known disease relevance. Proteins are annotated with the percentage of GO terms (with at least 10 human proteins) and high PScore-enriched GO terms (Panther analysis, PScore ≥4, with O/E > 1) of which the protein is a member, as well as the total number of each for which the annotated protein has the highest PScore in the set. Examples are grouped by (A), involvement in synaptic plasticity and neuronal behavior, showing synaptic functional regulator FMR1, and synaptophysin; (B), intracellular biomaterials and related structural proteins, showing focal adhesion kinase 1, vimentin, and keratin type I cytoskeletal 10; (C), proteins involved in signaling pathways, showing CCR4-NOT transcription complex subunit 3, β-catenin, vitamin D3 receptor, and Smoothened homolog; and (D), proteins involved in extracellular biomaterials, showing fibrinogen alpha chain and dentin sialophosphoprotein. (E) The cystic fibrosis transmembrane conductance regulator is shown as an example of a negative prediction, even though containing a large region of intrinsic disorder (residues ~650–840).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | His-SUMO-Ddx4 1-236 | PMID 25747659 | Expression vector (His-Sumo tagged) for Ddx4 residues 1–236, sequence from UID: Q9NQI0-1 (uniprot identification) | |
| Recombinant DNA reagent | His-SUMO-Ddx4 1-236(9FtoA) | PMID 25747659 | Expression vector (His-Sumo tagged) for Ddx4 residues 1–236, sequence from UID: Q9NQI0-1, 9 out of 14 phenylalanines mutated to alanine | |
| Recombinant DNA reagent | His-SUMO-Ddx4 1-236(14FtoA) | PMID 28894006 | Expression vector (His-Sumo tagged) for Ddx4 residues 1–236, sequence from UID: Q9NQI0-1, all phenylalanines mutated to alanine | |
| Recombinant DNA reagent | His-SUMO-Ddx4 1-236(RtoK) | PMID 28894006 | Expression vector (His-Sumo tagged) for Ddx4 residues 1–236, sequence from UID: Q9NQI0-1, all arginines mutated to lysine | |
| Recombinant DNA reagent | His-SUMO-FMR1445-632 | This paper | Expression vector (His-Sumo tagged) for FMR1 residues 445–632, sequence from UID: Q06787-1 | |
| Recombinant DNA reagent | His-SUMO-FMR1445-632(RtoK) | This paper | Expression vector (His-Sumo tagged) for FMR1 residues 445–632, sequence from UID: Q06787-1, all arginines mutated to lysine | |
| Recombinant DNA reagent | His-SUMO-pAPA341Q | This paper | Expression vector (His-Sumo tagged) for SCAF isoform pAP, sequence from UID: P16753-2, alanine 341 mutated to glutamine | |
| Recombinant DNA reagent | His-SUMO-EN2 | This paper | Expression vector (His-Sumo tagged) for Engrailed-2, sequence from UID: P19622-1 |
Contact statistics in high resolution, low R-factor protein structures.
https://doi.org/10.7554/eLife.31486.034| Measurement | Value | N | |||
|---|---|---|---|---|---|
| Contacts per 100 residues | |||||
| Pi-Contacts per 100 residues, averaged over PDBs | 6.06 ± 2.5* | 5,718 PDBs | |||
| Pi-Contacts per 100 residues, averaged over all residues | 6.27 ± 0.03 | 1,384,228 residues | |||
| Atom Contact Probabilities (%) | |||||
| Heavy Atoms in a Pi-Contact | 6.10 ± 0.03 | 10,836,487 atoms | |||
| sp2 Heavy Atoms in a Pi-Contact | 10.52 ± 0.05 | 6,283,150 atoms | |||
| Heavy Atoms within 4.9 Å of any Pi-Contact | 32.1 ± 0.1 | 10,836,487 atoms | |||
| Sidechain-Sidechain Contact Proportions (%) | 25,930 contacts | ||||
| Aromatic to Aromatic | 24.73 ± 0.29 | “ | |||
| Aromatic to Non-Aromatic | 53.24 ± 0.33 | “ | |||
| Non-Aromatic to Non-Aromatic | 22.03 ± 0.28 | “ | |||
| All Contact Proportions (%) | 86,860 contacts | ||||
| Sidechain to Sidechain | 29.85 ± 0.17 | “ | |||
| Aromatic Sidechain to Backbone | 40.41 ± 0.20 | “ | |||
| Non-Aromatic Sidechain to Backbone | 22.80 ± 0.16 | “ | |||
| Backbone to Backbone | 6.94 ± 0.09 | “ | |||
| Aromatic to Aromatic | 7.38 ± 0.10 | “ | |||
| outnumbered by Aromatic to Non-Aromatic | 7.6 ± 0.1 to 1 | “ | |||
| outnumbered by Non-Aromatic to Non-Aromatic | 3.9 ± 0.1 to 1 | “ | |||
| Arginine Sidechain Contacts (per 100 residues) | 61,877 residues | ||||
| Contact to Aromatic | 9.74 ± 0.13 | “ | |||
| Contact to Backbone | 10.6 ± 0.13 | “ | |||
| Contact to Glutamine/Asparagine Sidechain | 1.96 ± 0.06 | “ | |||
| Contact to Glutamate/Aspartate Sidechain | 1.49 ± 0.05 | “ | |||
| Contact to Arginine Sidechain | 3.63 ± 0.11 | “ | |||
-
*This error range shows the standard deviation between PDBs; other error ranges show standard error of the mean for averages computed over all PDBs.
Small molecule contact frequencies.
https://doi.org/10.7554/eLife.31486.035| Amino acid sp2group* | # PDBs | # Ligand Groups | Ligand Pi-Pi Contact Frequency (%) | # Protein Groups | Protein contact frequency (%) | O/E (Ligand/Protein) |
|---|---|---|---|---|---|---|
| GLU Sidechain | 84 | 209 | 16.3 ± 4.1 | 5353 | 5.0 ± 0.4 | 3.3 ± 0.8 |
| HIS Sidechain | 36 | 80 | 42.5 ± 8.0 | 530 | 17.4 ± 2.7 | 2.4 ± 0.7 |
| PHE Sidechain | 36 | 93 | 53.8 ± 9.0 | 1389 | 28.4 ± 2.3 | 1.9 ± 0.3 |
| ARG Sidechain | 61 | 145 | 43.9 ± 6.0 | 2878 | 15.9 ± 0.9 | 2.8 ± 0.5 |
| TYR Sidechain | 30 | 68 | 58.8 ± 12.2 | 806 | 19.9 ± 1.9 | 3.0 ± 0.7 |
| GLN Sidechain | 21 | 50 | 22.0 ± 8.9 | 800 | 13.0 ± 2.3 | 1.8 ± 0.8 |
| ASP Sidechain | 39 | 86 | 3.5 ± 2.0 | 2153 | 4.5 ± 0.8 | 0.8 ± 0.5 |
| TRP Sidechain | 43 | 109 | 50.5 ± 8.0 | 377 | 28.9 ± 2.3 | 1.7 ± 0.3 |
| ASN Sidechain | 11 | 32 | 6.3 ± 7.3 | 466 | 8.6 ± 1.9 | 0.7 ± 0.9 |
| Amino Carboxyl | 688 | 1704 | 8.9 ± 1.1 | 976 | 5.7 ± 1.2 | 1.5 ± 0.4 |
| Small molecule† | # PDBs | # Free Ligands | Ligand Pi-Pi Contact Frequency (%) | # sp2 Atoms | RCSB Ligand ID | Isomeric SMILES |
| Ethanal | 44 | 76 | 3.9 ± 2.9 | 2 | ACE | CC = O |
| Formic Acid | 444 | 2093 | 11.0 ± 0.8 | 3 | FMT | OC = O |
| Acetate Ion | 1664 | 4794 | 12.9 ± 0.6 | 3 | ACT | CC([O-])=O |
| Acetic Acid | 403 | 1133 | 13.5 ± 1.5 | 3 | ACY | CC(O)=O |
| Nitrate Ion | 225 | 852 | 15.3 ± 1.7 | 4 | NO3 | [O-][N+]([O-])=O |
| Guanidine | 32 | 115 | 15.7 ± 4.7 | 4 | GAI | NC(N)=N |
| Urea | 23 | 91 | 16.5 ± 4.0 | 4 | URE | NC(N)=O |
| Imidazole | 279 | 684 | 26.6 ± 2.4 | 5 | IMD | C1C[NH+]C[NH]1 |
-
*Entries containing amino acids or small sp2 containing planar molecules as free ligands were downloaded from the PDB (filtered to maximum sequence redundancy of 90% and 3 Å resolution) and pi-pi contact frequencies for ligands and their corresponding protein based equivalents were determined.
The majority of amino acids are more likely to form pi-pi contacts to protein when found as non-covalently bound ligands, rather than as residues within a protein, confirming that pi-pi contacts are a consistent property of amino acid interactions involving protein.
-
†In order to avoid bias due to the constrained geometries of functional binding sites we also analyzed the contact frequencies of a variety of common buffer components, with contact frequencies found to increase with number of sp2-hybridized atoms.
Ranges show standard error of the mean.
Pi-pi contact enrichment for catalytic residues.
Frequency of involvement in contacts, at either backbone or sidechain sp2 groups, is shown for individual residue types, residue independent (ANY), and residue type normalized (AVG), where catalytic residue contact frequency shows values for residues annotated as catalytic in the catalytic site atlas (Furnham et al., 2014) and non-catalytic residue contact frequency shows values for all other residues in the same structures. To normalize for possible differences in the number of contacts made by catalytic residues we also show number of pi-pi contacts divided by total number of VDW contacts, labeled as percent of VDW, and the percent of VDW ratio shows enrichment by dividing the catalytic percent of VDW value by the non-catalytic value. Error values are obtained by our standard bootstrap analysis (see Materials and methods), and enrichment values of greater than two standard deviations are shown in bold.
| Residue type | Non-catalytic contact frequency (%) | Catalytic residue contact frequency (%) | N catalytic | Enrichment | Non-catalytic percent of VDW (%) | Catalytic residue percent of VDW (%) | Percent of VDW ratio (cat./non) |
|---|---|---|---|---|---|---|---|
| ANY | 13.1 ± 0.1 | 24.5 ± 0.9 | 2914 | 1.87 ± 0.07 | 1.91 ± 0.09 | 3.94 ± 0.40 | 2.06 ± 0.23 |
| HIS | 31.9 ± 0.9 | 35.9 ± 2.3 | 471 | 1.12 ± 0.06 | 3.01 ± 0.26 | 4.33 ± 0.84 | 1.45 ± 0.31 |
| ASP | 12.9 ± 0.5 | 21.2 ± 2.0 | 448 | 1.65 ± 0.14 | 1.71 ± 0.22 | 3.08 ± 0.81 | 1.83 ± 0.54 |
| GLU | 11.7 ± 0.4 | 32.2 ± 2.5 | 370 | 2.75 ± 0.18 | 2.44 ± 0.31 | 6.57 ± 1.38 | 2.74 ± 0.71 |
| ARG | 26.7 ± 0.8 | 30.3 ± 3.2 | 287 | 1.14 ± 0.12 | 1.77 ± 0.29 | 7.85 ± 1.48 | 4.57 ± 1.26 |
| LYS | 6.2 ± 0.4 | 9.7 ± 2.0 | 259 | 1.56 ± 0.35 | 0.82 ± 0.20 | 0.37 ± 0.37 | 0.48 ± 0.52 |
| TYR | 36.1 ± 1.2 | 30.4 ± 3.7 | 171 | 0.84 ± 0.10 | 2.69 ± 0.39 | 4.62 ± 1.38 | 1.75 ± 0.59 |
| SER | 8.8 ± 0.5 | 13.0 ± 2.6 | 169 | 1.48 ± 0.28 | 0.83 ± 0.23 | 1.39 ± 0.70 | 1.84 ± 1.20 |
| CYS | 8.5 ± 1.3 | 14.7 ± 2.9 | 150 | 1.73 ± 0.26 | 1.45 ± 0.33 | 0.92 ± 0.66 | 0.68 ± 0.54 |
| ASN | 18.9 ± 1.1 | 26.6 ± 4.3 | 109 | 1.41 ± 0.20 | 1.88 ± 0.42 | 6.11 ± 1.76 | 3.42 ± 1.29 |
| GLY | 12.0 ± 0.7 | 16.2 ± 4.5 | 99 | 1.35 ± 0.41 | 1.26 ± 0.48 | 5.87 ± 1.92 | 5.72 ± 4.33 |
| THR | 6.8 ± 0.6 | 4.7 ± 2.3 | 86 | 0.69 ± 0.38 | 0.34 ± 0.18 | 0.88 ± 0.87 | 3.42 ± 4.17 |
| GLN | 18.0 ± 1.6 | 40.3 ± 7.3 | 62 | 2.24 ± 0.34 | 3.12 ± 0.53 | 1.88 ± 1.30 | 0.61 ± 0.44 |
| ALA | 7.8 ± 0.9 | 7.0 ± 3.3 | 57 | 0.91 ± 0.48 | 0.85 ± 0.43 | 1.28 ± 1.28 | 2.04 ± 2.75 |
| PHE | 34.2 ± 2.1 | 35.9 ± 7.1 | 53 | 1.05 ± 0.23 | 2.52 ± 0.64 | 4.92 ± 2.39 | 2.10 ± 1.27 |
| TRP | 46.2 ± 3.2 | 45.1 ± 7.5 | 51 | 0.98 ± 0.14 | 3.57 ± 0.73 | 2.88 ± 2.00 | 0.87 ± 0.68 |
| AVG | 18.7 ± 0.4 | 24.2 ± 1.2 | N/A | 1.42 ± 0.07 |
Effect of sp2 sidechain mutations on phase separation.
Phase separation critical concentration values for the N-terminus (1-236) of human Ddx4 and three mutants, 9FtoA and 14FtoA, as reported in (Nott et al., 2015), and RtoK, where all arginine residues have been mutated to lysine (Brady et al., 2017), as well as for the C-terminus (445-632) of human FMR1 and one mutant with all arginine residues mutated to lysine.
| Sample | Concentration at which phase separation is observed (conditions) | # F | # R | Total # | Mw (Da) |
|---|---|---|---|---|---|
| Ddx4 1–236 | (24°C, 20 mM Na2PO4 pH 6.5, 100 mM NaCl) | ||||
| WT | ~2 mg/mL | 14 | 24 | 236 | 25430 |
| 9FtoA | ~100 mg/mL | 5 | 24 | 236 | 24745 |
| 14FtoA | ~350 mg/mL | 0 | 24 | 236 | 24364 |
| RtoK | Not observed up to 400 mg/mL | 14 | 0 | 236 | 24758 |
| FMR1 445–632 | (4°C, 20 mM Na2PO4 pH 7.4, 2 mM DTT) | ||||
| WT | ~16 mg/mL | 2 | 28 | 188 | 20573 |
| RtoK | Not observed up to 216 mg/mL | 2 | 0 | 188 | 19789 |
Comparison of phase separation prediction and disorder prediction.
Two disorder predictors were tested on matched positive and negative sets to the phase separation predictor, comparing the relative discrimination of known phase-separating and known disordered proteins from the PDB, the human proteome, and the same set of known disordered proteins. AUC values are highlighted in blue for AUC >0.8, and red for AUC <0.7. Error values were obtained by bootstrap analysis.
| Positive set | AUC (vs. PDB) | AUC (vs. Human) | AUC (vs. Disprot) |
|---|---|---|---|
| Disopred3 (Disorder Predictor) | |||
| Phase Separation Test Set | 0.982 ± 0.005 | 0.72 ± 0.03 | |
| Disprot Set | 0.977 ± 0.007 | N/A | |
| IUPRED-Long (Disorder Predictor) | |||
| Phase Separation Test Set | 0.893 ± 0.007 | 0.70 ± 0.03 | |
| Disprot Set | 0.89 ± 0.01 | N/A | |
| PScore (Phase Separation Predictor) | |||
| Phase Separation Test Set | 0.961 ± 0.005 | 0.88 ± 0.01 | 0.84 ± 0.01 |
| Disprot Set | 0.79 ± 0.02 | N/A | |
Retrospective analysis of predictor quality at different stages during the training process.
AUC values for distinguishing proteomic phase-separating sequences from the human proteome are shown for prediction scores made from pi-contact frequencies (average contacts predicted per residue) obtained at each training step of the protocol in order of their sequential development, with prediction scores calculated as the highest number of contacts predicted for any given 100 residue window in each sequence. Analysis of the relative effects of different contact types was added by excluding contacts from each score and retesting. Standard error of the mean (SEM), by bootstrap analysis, is consistently in the range from 0.021 to 0.039.
| Training step | AUC at training step | Sidechain contacts only | Backbone contacts only | Short-range sidechain only | Long-range sidechain only | Short-range backbone only | Long-range backbone only |
|---|---|---|---|---|---|---|---|
| (1) Baseline Frequencies | 0.57 | 0.51 | 0.84 | 0.52 | 0.50 | 0.73 | 0.80 |
| 2) Context-Averaged Frequencies | 0.57 | 0.51 | 0.86 | 0.53 | 0.51 | 0.77 | 0.83 |
| (3) Smoothed Frequency Predictions | 0.82 | 0.64 | 0.89 | 0.59 | 0.65 | 0.71 | 0.85 |
| (4) Weight Optimized Final Predictor | 0.88 | N/A | N/A | N/A | N/A | N/A | N/A |
Sequence similarity comparison.
Frequencies of dipeptides (pairs of neighboring amino residues) were computed for phase-separating proteins and the human proteome, and enrichment was measured by the percentage of human proteins with lower frequency than found in a given sequence. The fifteen dipeptides enriched (≥99%) in the most sequences within the phase separation test sets are shown in the table vs. enrichment values obtained for the phase separation training set and three experimentally verified proteins. Values in the top fifth percentile are shown in bold.
| Protein Name | Dipeptide enrichment (Percentage of human proteome with lower frequency) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GV | VG | VP | PG | FG | RG | GR | GG | YG | GS | SG | GA | GF | GD | DS | |
| Training Set Proteins | |||||||||||||||
| Elastin | 100 | 100 | 100 | 100 | 97 | 31 | 32 | 99 | 99 | 20 | 20 | 100 | 89 | 38 | 30 |
| Nsp1 | 30 | 34 | 31 | 26 | 100 | 31 | 30 | 75 | 52 | 90 | 38 | 99 | 60 | 66 | 68 |
| TIA1 | 73 | 75 | 46 | 26 | 86 | 31 | 86 | 77 | 99 | 29 | 53 | 26 | 84 | 54 | 30 |
| LAF1 | 30 | 78 | 65 | 29 | 67 | 99 | 99 | 100 | 77 | 88 | 97 | 65 | 78 | 97 | 32 |
| EIF4H | 30 | 65 | 31 | 52 | 98 | 99 | 95 | 99 | 52 | 99 | 42 | 79 | 99 | 98 | 89 |
| Ddx3x | 51 | 70 | 34 | 43 | 89 | 98 | 97 | 96 | 93 | 93 | 95 | 68 | 96 | 59 | 78 |
| hnRNPA1 | 30 | 55 | 31 | 44 | 100 | 99 | 99 | 100 | 99 | 99 | 98 | 44 | 99 | 60 | 79 |
| DDX4 | 33 | 77 | 48 | 53 | 98 | 96 | 91 | 89 | 59 | 87 | 96 | 29 | 98 | 96 | 45 |
| FUS | 30 | 31 | 31 | 83 | 78 | 100 | 99 | 100 | 100 | 98 | 99 | 33 | 93 | 91 | 57 |
| EWS | 52 | 31 | 35 | 97 | 51 | 100 | 99 | 100 | 100 | 72 | 61 | 30 | 97 | 91 | 48 |
| TAF15 | 36 | 38 | 31 | 30 | 53 | 100 | 99 | 100 | 100 | 92 | 99 | 26 | 71 | 100 | 94 |
| Experimentally Verified Proteins | |||||||||||||||
| FMR1 | 69 | 89 | 93 | 44 | 43 | 96 | 94 | 83 | 62 | 62 | 34 | 70 | 48 | 41 | 67 |
| SCAF pAP | 75 | 50 | 89 | 91 | 43 | 49 | 73 | 97 | 75 | 92 | 96 | 96 | 40 | 36 | 44 |
| Engrailed-2 | 30 | 31 | 31 | 97 | 70 | 78 | 90 | 100 | 52 | 99 | 91 | 99 | 40 | 95 | 97 |
High PScore enrichment for human proteins with a greater than average number of post-translational modification (PTM) site annotations in Phosphosite+.
PTM counts are controlled for protein length by taking the maximum number observed in any 100 residue window, and the threshold for an above average PTM count is defined as greater than the average plus one standard deviation. Errors show SEM by bootstrap analysis.
| Phosphosite+ PTM annotation type | PTM count threshold | Above threshold (N) | PScore > 4 (%) | Enrichment |
|---|---|---|---|---|
| O-GlcNAc | 1 | 158 | 17 ± 3 | 3.4 |
| Methyl | 2 | 2051 | 13.3 ± 0.7 | 2.7 |
| Phosphate | 10 | 2485 | 10.8 ± 0.8 | 2.2 |
| O-GalNAc | 1 | 456 | 10.1 ± 0.1 | 2.0 |
| Sumo | 1 | 1999 | 9.0 ± 0.7 | 1.8 |
| Acetyl | 3 | 1543 | 8.0 ± 0.7 | 1.6 |
| Ubiquitin | 4 | 1875 | 6.3 ± 0.5 | 1.3 |
| Disease Relevant | 1 | 298 | 11 ± 2 | 2.1 |
| Regulatory Function | 2 | 1087 | 7.6 ± 0.8 | 1.5 |
| Database Baseline | 0 | 18582 | 5.0 ± 0.2 | 1.0 |
Additional files
-
Source code 1
Python scripts for identifying PDB contacts.
Pi-pi contact identification scripts suitable for reproducing the annotation data contained in Figure 1—source data 1 and 2.
- https://doi.org/10.7554/eLife.31486.021
-
Source code 2
Final predictor code package.
Python script and associated database files for the final phase separation propensity predictor.
- https://doi.org/10.7554/eLife.31486.022
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31486.023





