Synthesizing artificial devices that redirect cellular information at will
Figures
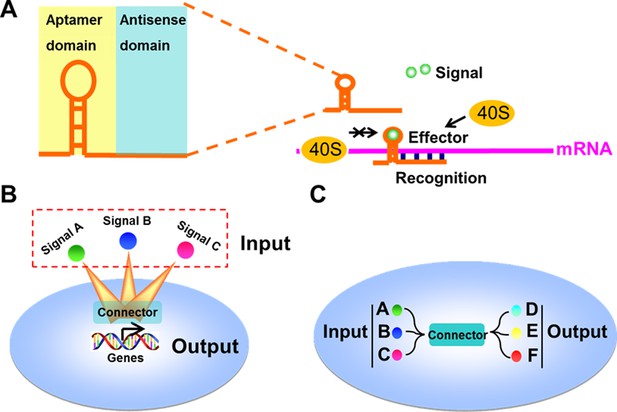
Design and construction of the signal-connectors for constructing linkages between signaling nodes.
(A) Domain composition of a signal-connector. The device uses the antisense domain to recognize the mRNA of a target gene and the aptamer domain to respond to different signals to control the translation of the target gene. (B) Connectors that control the expression of cellular genes in response to specific exogenous signals can be engineered through this modular approach. (C) The signal-connectors can be used to direct the linkages between cellular inputs and outputs.
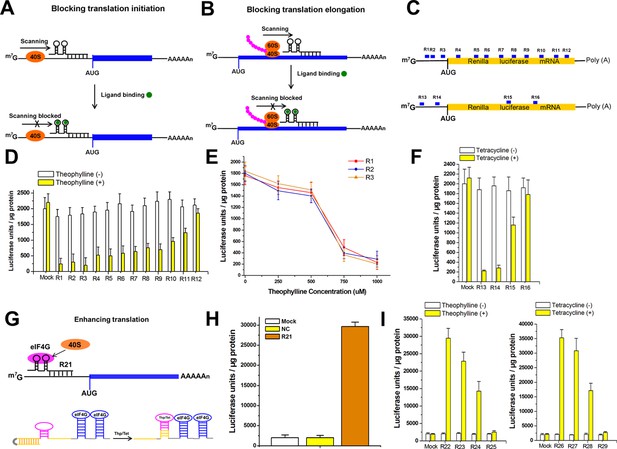
The signal-connectors effectively silence/activate gene expression.
(A) Design of the signal-connector to block translation of the targeted gene. The device is designed to bind the 5ʹ-UTR of the targeted mRNA. In the absence of the ligand, scanning of the 40S ribosome subunit proceeds until the AUG start codon is reached and translation is initiated. In the presence of the ligand, the ligand-aptamer complex disturbs ribosome scanning and blocks translation initiation. (B) When the binding occurs on the protein-coding region of target mRNA, it blocks ribosome and translation elongation. (C) Various signal-connectors were designed to target different regions of Renilla luciferase mRNA. (D) Renilla luciferase activity was suppressed by the signal-connectors only in the presence of 1 mM theophylline. Empty pGPU6/GFP/Puro vector as used as mock control. (E) Cells stably transfected with the signal-connectors (R1, R2 and R3) were grown in the presence of 0, 250, 500, 750, or 1000 µM theophylline. Addition of theophylline inhibited in vitro translation of the Renilla luciferase mRNA in a dose-dependent manner. (F) Suppression of Renilla luciferase in HEK293 cells by four different signal-connectors that bind to 100 µM tetracycline. (G) Design of the signal-connector for enhancing translation of a targeted gene. The activation of translation by a signal-connector is due to the activation of formation of initiation factor complexes involving eIF4G. In the absence of theophylline/tetracycline, the antisense domain is unable to bind to the mRNA of its target gene. In the presence of theophylline/tetracycline, the aptamer stem is formed and the antisense domain will bind to its target. (H) Renilla luciferase activity was increased by the signal-connector. (I) Addition of theophylline/tetracycline increased translation of the Renilla luciferase mRNA. Empty pGPU6/GFP/Puro vector as used as mock control. NC, negative control vector with two repeated elements not having targets in the human genome. Reported data are mean ± SD from at least three experiments.
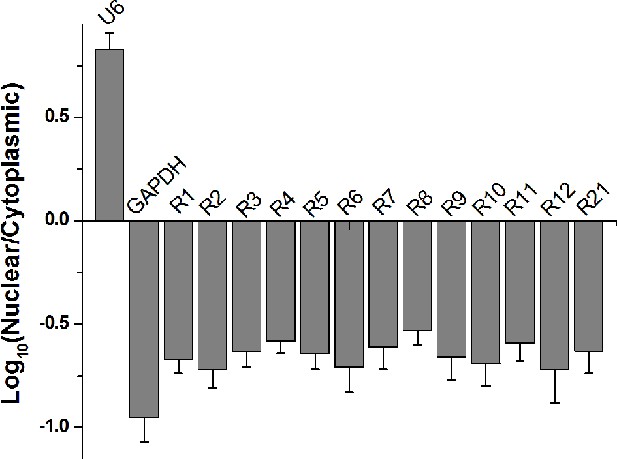
Nuclear and cytoplasmic RNA fractionation analysis.
The levels of signal-connectors in purified nuclear or cytoplasmic RNAs were detected using real-time qPCR. U6 and GAPDH were used as nuclear and cytoplasmic control, respectively. Data are shown as mean ± SD based on three independent experiments.
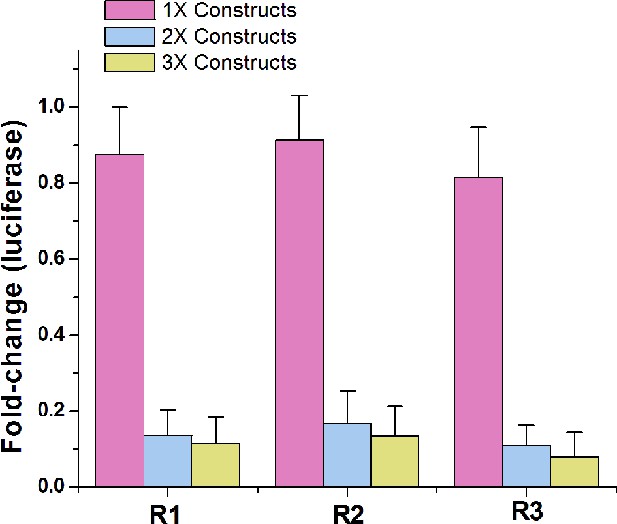
Comparing the repression effects of signal-connectors with different numbers of aptamers.
While 2 × devices and 3 × devices induced strong inhibitory effects on luciferase expression in the presence of 1 mM theophylline, the 1 × devices only caused minimal changes in luciferase expression. Fold change values are luciferase expression levels (mean ± SD) from three measurements, relative to negative control cell lines.
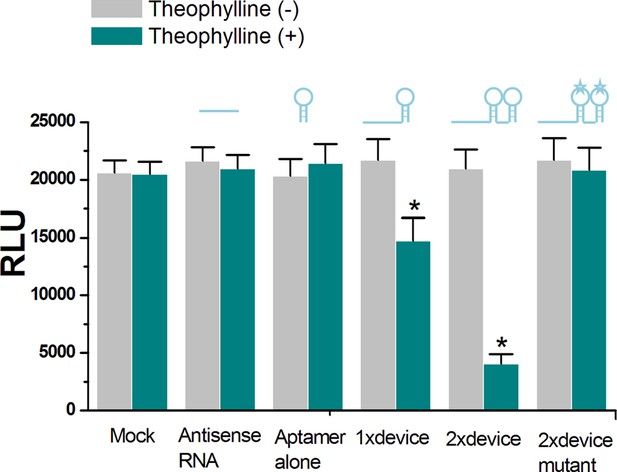
The in vitro translational repression activity induced by the ligand-aptamer complex.
The RNA ‘signal-connector’ (R1 used in Figure 2C of main text) resulted in decreased translation of the Renilla luciferase only in the presence of theophylline, as measured by luciferase activity. In addition, the 2 × signal connector (2 copies of aptamer) induced a higher reduction in translation of reporter gene than that of the 1 × signal connector (only one aptamer). The 2 × signal connector containing mutant theophylline aptamers could not reduce luciferase translation regardless of whether theophylline was present. Reported values are presented as mean ± SD and the experiments were repeated three times. *p<0.01.
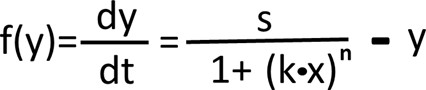
The mathematical equation for gene repression induced by the signal-connector.
The properties of the ‘OFF’ switch are explained by the above model, where y is the output, x is the concentration of the input signal, k is the number of the aptamers, n represents inhibitory parameter which is negatively correlated with the target distance from the 5’UTR, s is the normal expression rate of y in physiological status and –y represents the degradation rate of y in cell.
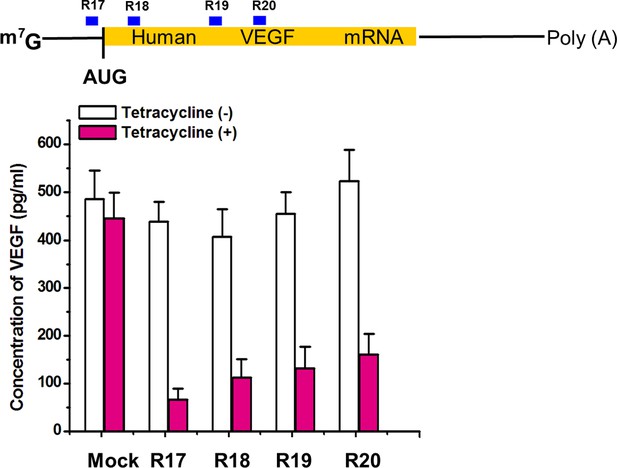
Suppression of VEGF protein expression in HEK293 cells by signal-connectors that bind to 100 µM tetracycline.
Four signal-connectors were designed to target different regions of human VEGF mRNA. Empty pGPU6/GFP/Puro vector as used as mock control. Reported values are presented as mean ± SD and the experiments were repeated three times.
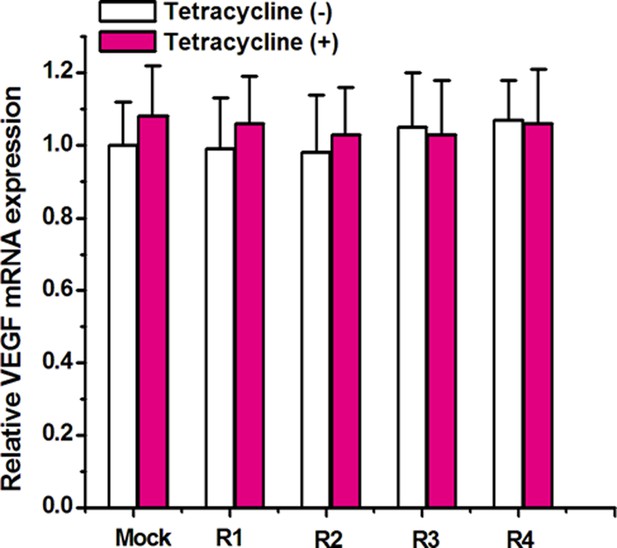
Relative expression level of VEGF mRNA.
Quantitative real-time PCR was performed to determine the expression level of VEGF mRNA. GAPDH was used as the internal control. Relative levels of VEGF mRNA did not change between cells harboring signal-connectors grown in the absence or presence of tetracycline. Reported values are presented as mean ± SD and the experiments were repeated at least three times. Mock, empty pGPU6/GFP/Puro vector.
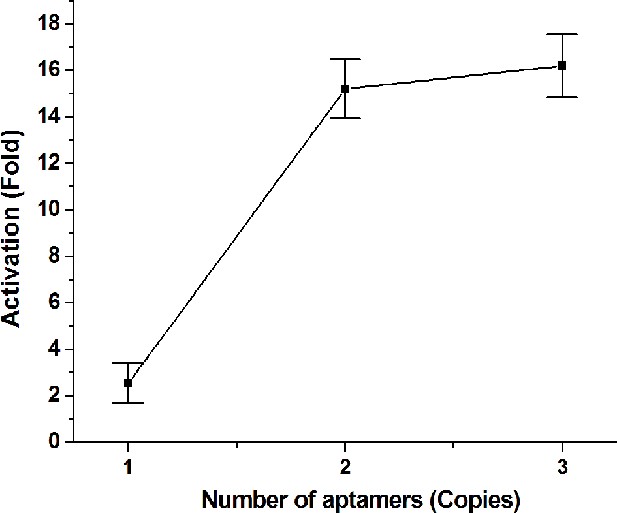
Comparing the activation effects of signal-connectors with different numbers of aptamers.
While 2 × device and 3 × device caused high activation fold changes on luciferase expression in the presence of 1 mM theophylline, the 1 × devices only induced minimal increase in luciferase expression. Fold change values are luciferase expression levels relative to negative control cell lines. Values are mean ± SD from three independent measurements.
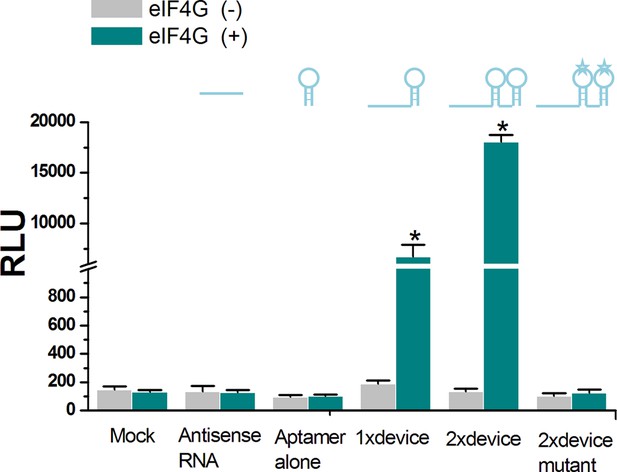
The in vitro translational activation activity induced by the ligand-aptamer complex.
Neither the single eIF4G aptamer nor the antisense domain alone induced in vitro luciferase translation regardless of whether eIF4G was present. Instead, the RNA ‘signal-connector’ (R21 used in Figure 2G of main text) resulted in translation of the Renilla luciferase in the presence of eIF4G, as measured by luciferase activity. In addition, the 2 × signal connector (2 copies of aptamer) induced a higher activation in translation of reporter gene than that of the 1 × signal connector (only 1 aptamer). The 2 × signal connector containing mutant eIF4G aptamers could not induce luciferase translation regardless of whether eIF4G was present. Reported values are presented as mean ± SD and the experiments were repeated three times.*p<0.01.

The mathematical equation for gene activation induced by the signal-connector.
The properties of the ‘ON’ switch are explained by the above model, where y is the output, x is the concentration of the input signal (eIF4G), k is the number of the aptamers, n represents activation parameter which is negatively correlated with the target distance from the 5’UTR, s is the normal expression rate of y in physiological status and –y represents the degradation rate of y in cell.
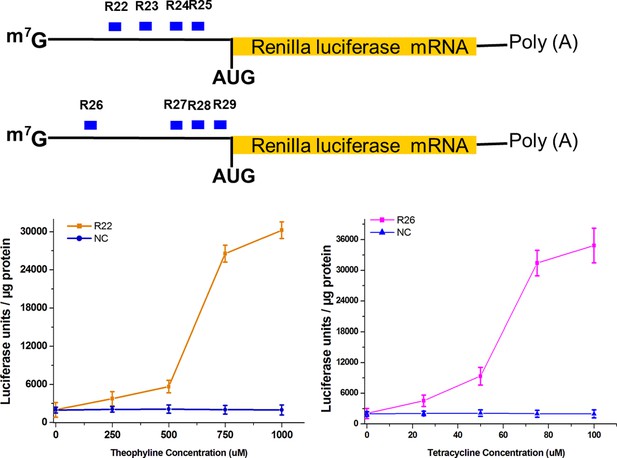
Addition of theophylline/tetracycline increased translation of the Renilla luciferase mRNA in a dose-dependent manner.
Empty pGPU6/GFP/Puro vector as used as mock control. NC, negative control vector with two repeated elements not having targets in the human genome. Reported data are mean ± SD from at least three experiments.
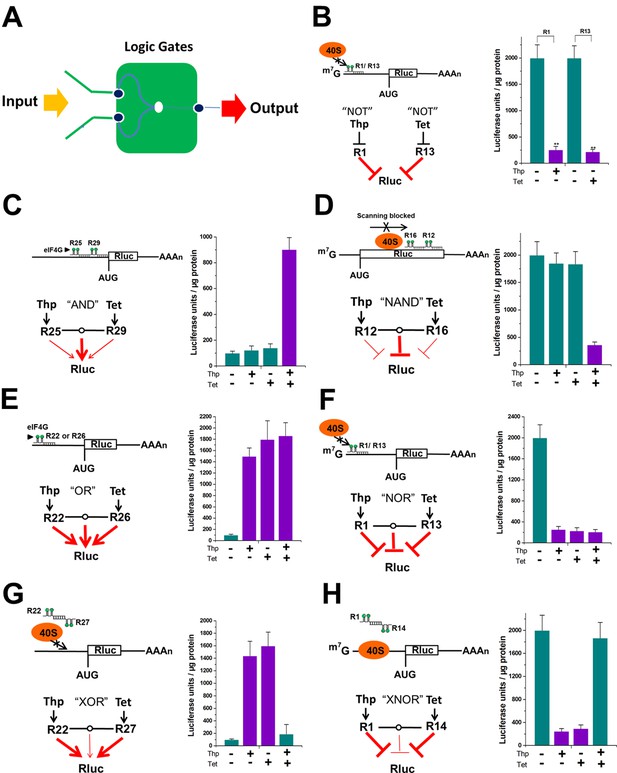
Logic gates based on signal integration were constructed by the signal-connectors.
(A) In each logic gate, the signal-connector integrates the input signals theophylline and tetracycline and produces a luciferase output. Construction of gates that perform NOT (B), AND (C), NAND (D), OR (E), NOR (F), XOR (G) and XNOR (H) functions using the signal-connectors.
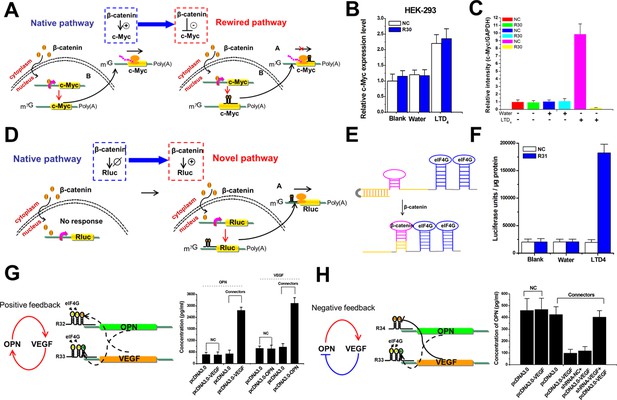
The signal-connectors effectively rewire and create signaling pathways and feedback loops.
(A) Mechanisms of the signal-connectors designed to rewire the signaling pathway. (B) The relative expression levels of c-Myc mRNA were evaluated using real-time qPCR in HEK293 cells. The level of c-Myc mRNA was increased in cells that respond to LTD4 stimulation. (C) Histogram of c-Myc. The values were normalized to GAPDH for each sample. The negative control was defined as 1.0. (D) Mechanisms of the signal-connectors designed to create the signaling pathway. (E) Design of the signal-connector that responds to β-catenin. This device frees the antisense region and targets the mRNA only in the presence of β-catenin. (F) The activity of Renilla luciferase was evaluated in HEK293 cells that respond to LTD4 stimulation. (G) Designed models and experimental results illustrating the putative roles of the signal-connectors in constructing the OPN–VEGF positive feedback loop. (H) Designed models and experimental results illustrating the putative roles of the signal-connectors in constructing the OPN–VEGF negative feedback loop. NC, negative control vector with two repeated elements not having targets in the human genome. Reported values are presented as mean ± SD and the experiments were repeated at least three times.
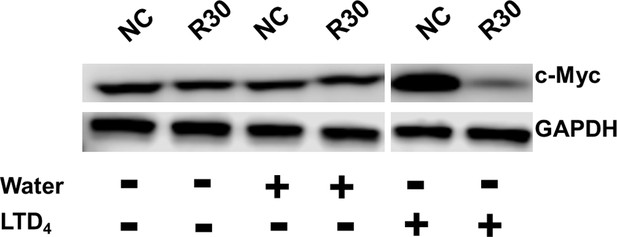
Representative images of western blot analysis of c-Myc protein expression in cells transfected with the signal-connector or the negative control (NC).
https://doi.org/10.7554/eLife.31936.017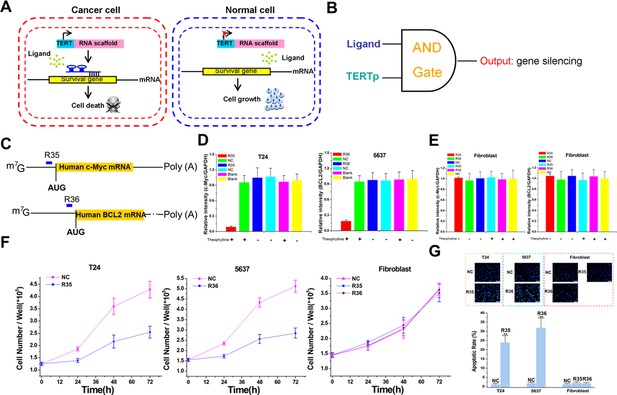
The signal-connectors specifically silence survival gene expression and inhibit cell growth in the targeted cancer cells.
(A) Mechanisms of the signal-connectors designed to selectively kill cancer cells, which control cell survival in response to the presence of activated TERTp and ligand. (B) A schematic representation of the genetic AND gate. hTERT promoter and ligand (1000 µM theophylline) are the two inputs of the circuit. (C) Two different signal-connectors were designed to target the 5ʹ-UTRs of human c-Myc mRNA and BCL2 mRNA. (D and E) Quantitative western blot analysis of targeted protein expression in bladder cancer cells (T24 and 5637) and normal fibroblast cells. NC, negative control vector with two repeated elements not having targets in the human genome. Blank, cells that were not transfected with vector. (F) Cell growth was measured by CCK-8 assay at different time intervals. ANOVA was used for the comparison of cell growth curves. Reported values are mean ± SD from three independent experiments.
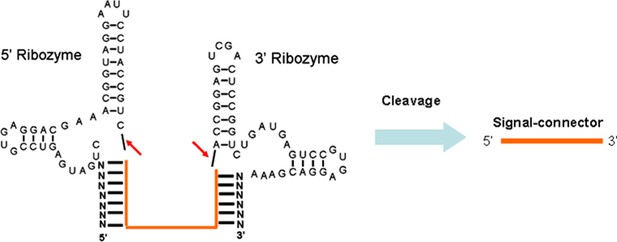
Sequence and cleavage mechanism of the ribozymes.
The designed signal-connector sequence is inserted between the two ribozyme sequences. The two cleavage sites are indicated (red small arrows).The sequence of signal-connector can be released from the primary RNA transcript due to action of the ribozymes and therefore be free of potentially interfering flanking sequences.
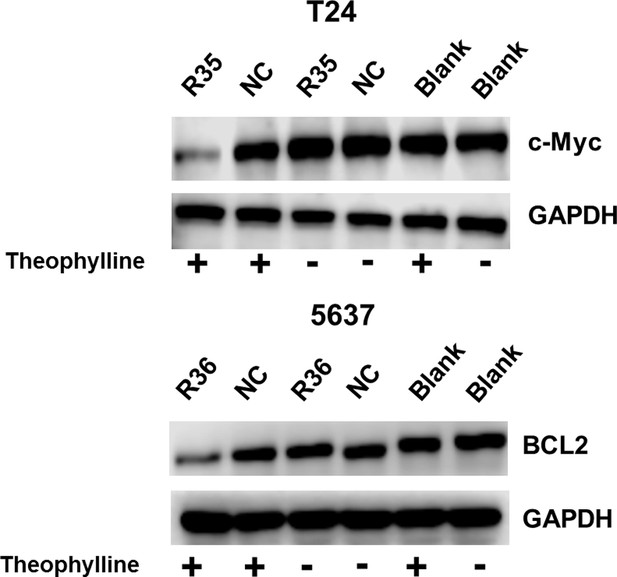
Representative images of western blot analysis of c-Myc/BCL2 protein expression in bladder cancer cells transfected with the signal-connector or the controls.
https://doi.org/10.7554/eLife.31936.020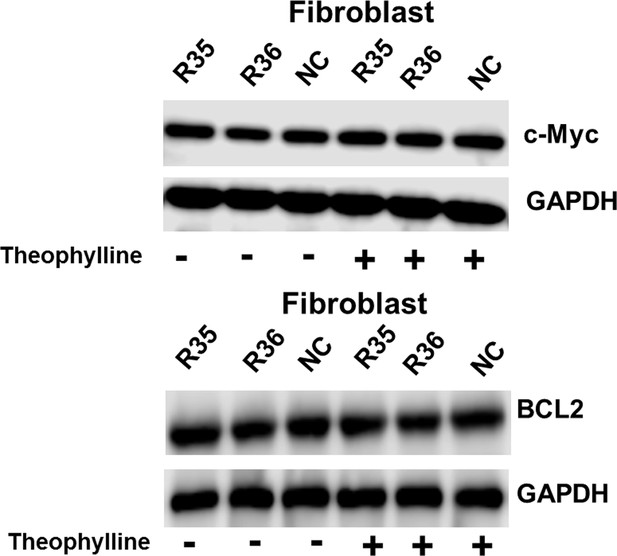
Representative images of western blot analysis of c-Myc/BCL2 protein expression in Fibroblast transfected with the signal-connector or the controls.
https://doi.org/10.7554/eLife.31936.021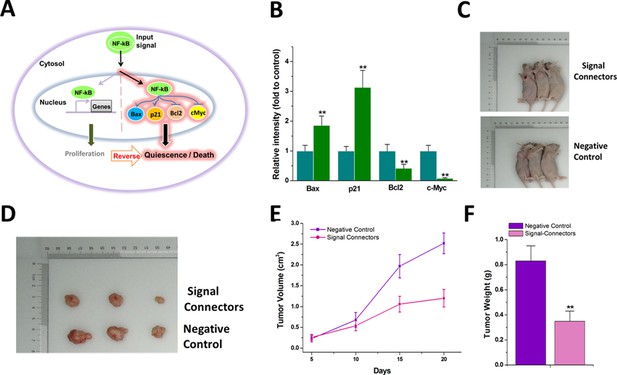
The signal-connector induces simultaneous activation and repression of cellular genes in response to oncogenic signal and redirects the oncogenic signaling to an anti-oncogenic pathway.
(A) The oncogenic signal NF-kB was redirected to activate two tumor suppressors, Bax and p21, and to suppress two oncogenes, BCL2 and c-Myc, by the signal-connector in the cancer cells. (B) The relative expression levels of BAX, BCL2, c-Myc and p21 were determined in T24 cells by quantitative western blot at 48 hr after cell seeding. Reported values are mean ± SD from three independent experiments. (C and D) 20 days after injection, tumors formed in the signal-connector group were dramatically smaller relative to negative control. (E) The tumor volume was calculated once every 5 days after injection of T24 cells stably transfected with signal-connector or negative control. Bars indicate SD. (F) Tumor weights are shown as means of tumor weight ± SD. **p<0.01.
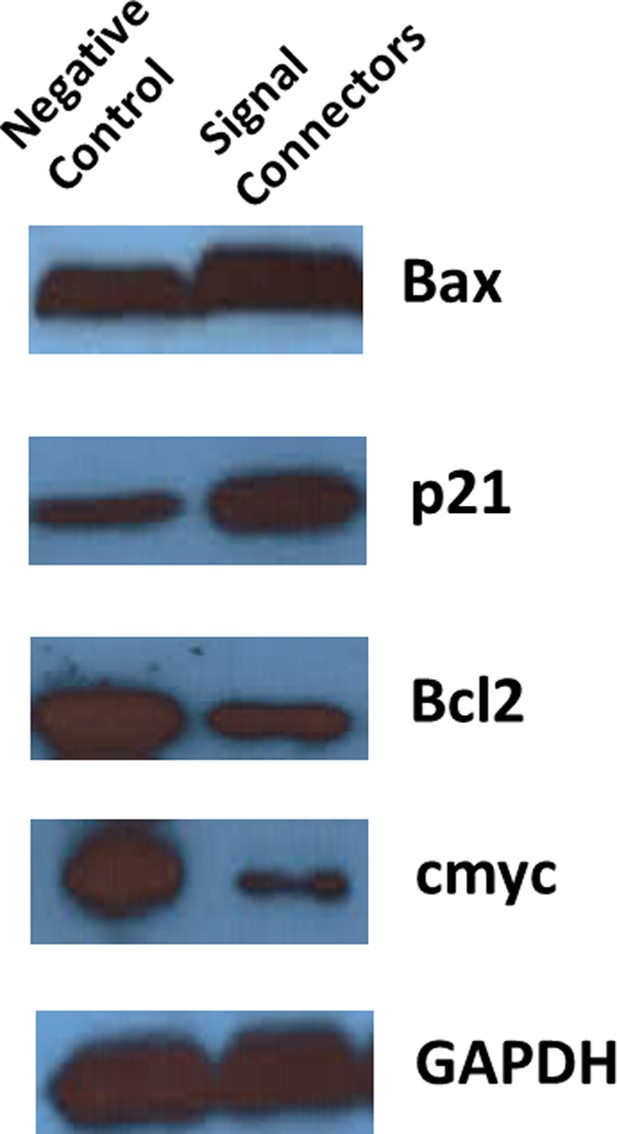
Representative images of western blot analysis of targeted protein expression in T24 cells transfected with the signal-connectors or the negative control.
https://doi.org/10.7554/eLife.31936.023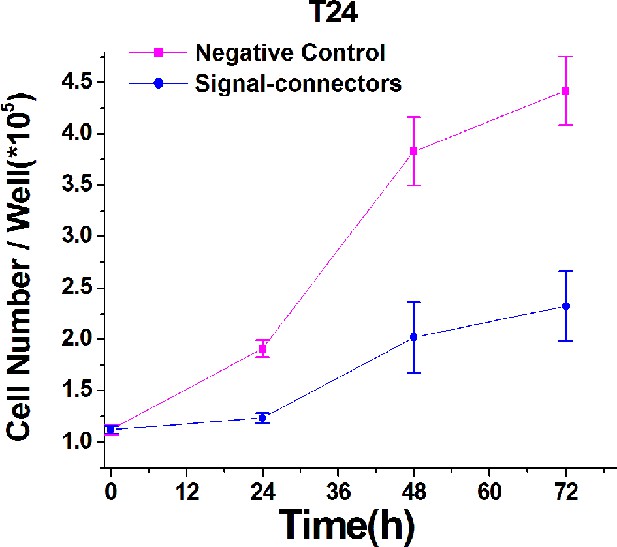
Growth curves of T24 cell lines transfected with either the signal-connectors or the negative control vector.
Proliferations of the transfected T24 cells were measured by CCK-8 assay at different time intervals. The curves of cell proliferation were compared using ANOVA. Result at each time point is shown as mean ±S.D.
Additional files
-
Supplementary file 1
cDNA sequences of the theophylline-induced signal-connectors targeting and suppressing Renilla luciferase mRNA translation.
Each of these sequences consists of a complementary sequence, two copies of theophylline aptamers, and a linker sequence.
- https://doi.org/10.7554/eLife.31936.025
-
Supplementary file 2
cDNA sequences of the tetracycline-induced signal-connectors targeting and suppressing Renilla luciferase mRNA translation.
Each of these sequences consists of a complementary sequence, two copies of tetracycline aptamers, and a linker sequence.
- https://doi.org/10.7554/eLife.31936.026
-
Supplementary file 3
cDNA sequences of the tetracycline-induced signal-connectors targeting and suppressing VEGF mRNA translation.
Each of these sequences consists of a complementary sequence, two copies of tetracycline aptamers, and a linker sequence.
- https://doi.org/10.7554/eLife.31936.027
-
Supplementary file 4
The cDNA sequence of the signal-connector targeting and enhancing Renilla luciferase mRNA translation.
The sequence consists of a complementary sequence, two copies of eIF4G aptamers, and a linker sequence.
- https://doi.org/10.7554/eLife.31936.028
-
Supplementary file 5
cDNA sequences of theophylline-induced signal-connectors targeting and enhancing Renilla luciferase mRNA translation.
Each of these sequences consists of a complementary sequence, one copy of theophylline riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.029
-
Supplementary file 6
cDNA sequences of tetracycline-induced signal-connectors targeting and enhancing Renilla luciferase mRNA translation.
Each of these sequences consists of a complementary sequence, one copy of tetracycline riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.030
-
Supplementary file 7
The cDNA sequence of β-catenin-induced signal-connector targeting and suppressing c-Myc mRNA translation.
The sequence consists of a complementary sequence, two copies of β-catenin aptamers, and a linker sequence.
- https://doi.org/10.7554/eLife.31936.031
-
Supplementary file 8
The cDNA sequence of β-catenin-induced signal-connector targeting and enhancing Renilla luciferase mRNA translation.
The sequence consists of a complementary sequence, one copy of β-catenin riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.032
-
Supplementary file 9
The cDNA sequence of VEGF-induced signal-connector targeting and enhancing OPN mRNA translation.
The sequence consists of a complementary sequence, one copy of VEGF riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.033
-
Supplementary file 10
The cDNA sequence of OPN-induced signal-connector targeting and enhancing VEGF mRNA translation.
The sequence consists of a complementary sequence, one copy of OPN riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.034
-
Supplementary file 11
The cDNA sequence of VEGF-induced signal-connector targeting and suppressing OPN mRNA translation.
The sequence consists of a complementary sequence, two copies of VEGF aptamers and one linker sequence.
- https://doi.org/10.7554/eLife.31936.035
-
Supplementary file 12
The cDNA sequence of theophylline-induced signal-connector targeting and suppressing c-Myc mRNA translation.
The sequence consists of a complementary sequence, two copies of theophylline aptamers and one linker sequence.
- https://doi.org/10.7554/eLife.31936.036
-
Supplementary file 13
The cDNA sequence of theophylline-induced signal-connector targeting and suppressing BCL2 mRNA translation.
The sequence consists of a complementary sequence, two copies of theophylline aptamers and one linker sequence.
- https://doi.org/10.7554/eLife.31936.037
-
Supplementary file 14
The cDNA sequence of NF-kB-induced signal-connector targeting and enhancing Bax mRNA translation.
The sequence consists of a complementary sequence, one copy of NF-kB riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.038
-
Supplementary file 15
The cDNA sequence of NF-kB-induced signal-connector targeting and enhancing p21 mRNA translation.
The sequence consists of a complementary sequence, one copy of NF-kB riboswitch, two copies of eIF4G aptamers and two linker sequences.
- https://doi.org/10.7554/eLife.31936.039
-
Supplementary file 16
The cDNA sequence of NF-kB-induced signal-connector targeting and suppressing BCL2 mRNA translation.
The sequence consists of a complementary sequence, two copies of NF-kB aptamers and one linker sequence.
- https://doi.org/10.7554/eLife.31936.040
-
Supplementary file 17
The cDNA sequence of NF-kB-induced signal-connector targeting and suppressing c-Myc mRNA translation.
The sequence consists of a complementary sequence, two copies of NF-kB aptamers and one linker sequence.
- https://doi.org/10.7554/eLife.31936.041
-
Transparent reporting form
- https://doi.org/10.7554/eLife.31936.042





