Natural changes in light interact with circadian regulation at promoters to control gene expression in cyanobacteria
Figures
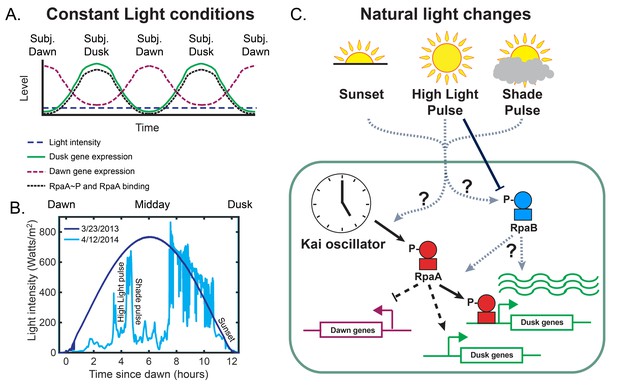
The circadian and light response pathways in cyanobacteria.
(A) Schematic of gene expression output of the circadian clock under Constant Light conditions. Under Constant Light intensity (dashed navy blue line), dawn gene expression (dashed maroon line) and dusk gene expression (solid green line) display oscillatory patterns, peaking at subjective dawn and subjective dusk, respectively. The Kai post-translational oscillator generates oscillations in the levels of phosphorylated RpaA (RpaAP) and the binding of RpaA to DNA (black dotted line), with the peak amplitude at subjective dusk. (B) Solar irradiance measurements in units of watts m at 342.5 meters above sea level in Madison, WI, on a clear day (3/23/13, dark blue), and a day on which fluctuations in cloud cover generated rapid changes in light intensity (4/12/14, light blue) (Petty and Weidner, 2017). Examples of a ‘High Light pulse,’ ‘Shade pulse,’ and ‘Sunset’ are indicated. (C) Schematic of the regulation of circadian gene expression. RpaA phosphorylation state converts timing information from the Kai oscillator to changes in gene expression by directly binding and activating a subset of dusk genes, indirectly activating the remainder of the dusk genes, and indirectly repressing the dawn genes. High Light Pulse conditions cause dephosphorylation of RpaB (Moronta-Barrios et al., 2012), but the effects of conditions like Sunset or Shade on RpaB are unknown. It is unclear whether natural fluctuations in light directly affect the clock and its output pathways and how light-induced changes in RpaB activity might be involved.
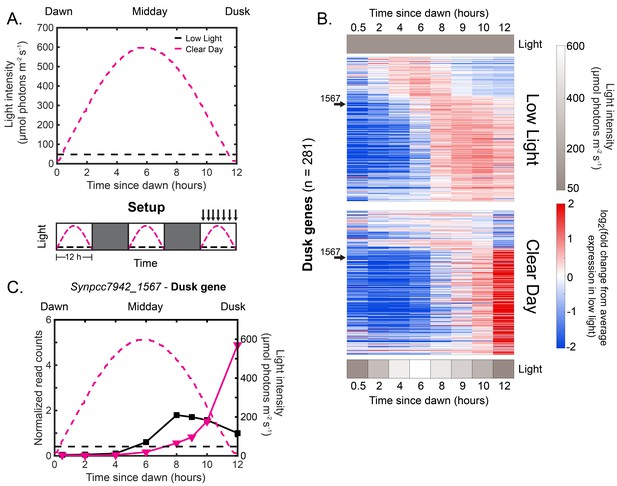
Natural clear day conditions sharpen the expression of dusk genes to peak just before expected darkness.
(A) Experimental setup for testing the effects of Clear Day conditions on circadian gene expression. The upper plot shows the light intensity profiles of Low Light (black) and Clear Day (magenta) conditions, in units of mol photons m s (see Materials and methods - Calibrating light conditions for more details; light intensity values available in Figure 2—source data 1). The lower plot displays the experimental setup. Cells were grown under Clear Day (magenta dashed lines) or Low Light conditions (black dashed lines) for 12 hr, followed by 12 hr of darkness (dark gray boxes) for three days, with sampling over the third light period (indicated by arrows above plot). (B) Gene expression dynamics of all dusk genes (=281) under Low Light (top) and Clear Day (bottom) conditions. Gene expression is quantified as the log fold change from the average expression of the gene over all time points in the Low Light condition (see Materials and methods - RNA sequencing for more details; data available in Figure 2—source data 1). Genes were sorted by phase under Constant Light conditions (Vijayan et al., 2009). Light intensity at each time point is indicated in a grayscale heat map next to the corresponding condition. The data for a representative dusk gene, Synpcc7942_1567, is indicated with arrows. (C) Gene expression dynamics of the representative dusk gene Synpcc7942_1567 under Low Light (black) and Clear Day (magenta) conditions (left y-axis). The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis.
-
Figure 2—source data 1
Normalized gene expression in Low Light and Clear Day conditions.
- https://doi.org/10.7554/eLife.32032.009

Pigment levels of cyanobacteria grown under Low Light or Clear Day conditions reveal adjustments in the photosynthetic apparatus to optimize growth in different light conditions.
(A) Estimation of phycocyanin and chlorophyll levels in cells grown under Low Light (black) or Clear Day (magenta) conditions for two days, measured at midday of the third light period. Phycocyanin and chlorophyll levels were estimated by measuring optical density of the culture at 620 nm or 680 nm, respectively, and normalizing to optical density at 750 nm to account for differences in cell density. Error bars show the standard deviation of three independent measurements. Cells grown under Clear Day conditions show lower levels of both phycocyanin and chlorophyll. (B) Image of cells harvested from cultures at midday on the third day (OD = 0.3) following two 12 hr light/12 hr dark cycles in either Low Light (left) and Clear Day (right) conditions (see Figure 2A). The more yellow-green color of the cells from Clear Day conditions compared to the Low Light grown cells, which are blue-green, indicates diminished levels of phycocyanin. Cells grown in Clear Day divided roughly twice as fast at midday compared to Low Light cells (6 hr doubling in Clear Day compared to 12 hr doubling time in Low Light). The growth rate of cells grown in Clear Day conditions varied throughout the day, showing slowest growth just prior to dusk (10 hr doubling time). Growth rate was estimated by calculating the change in the of the culture before and after a 1 hr time period. Taken together, the growth and pigment differences suggest that the cells lowered the levels of photosynthetic components to cope with the increased photon flux of Clear Day, while using these additional resources for faster growth.
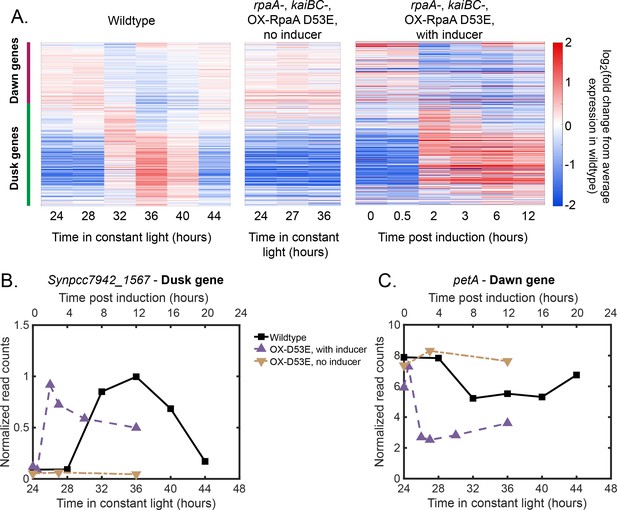
Gene expression dynamics of dusk and dawn circadian genes under Constant Light conditions (data from Markson et al., 2013).
(A) Gene expression dynamics of circadian genes over 24 hr in Constant Light conditions in wildtype cells (left heat map) and over 12 hr in OX-D53E cells (rpaA-, kaiBC-, Ptrc::rpaA(D53E)) (middle and right heat maps) as measured by RNA sequencing. The OX-D53E strain allows experimental control of RpaA activity via IPTG-inducible expression of the RpaA phosphomimetic RpaA-D53E in cells that lack wildtype RpaA (Markson et al., 2013). In the middle panel RpaA-D53E is not induced, and in the right panel IPTG was added to induce RpaA-D53E. Gene expression is quantified as the log fold change from the average expression of the gene over all time points in the wildtype cells under the Constant Light condition. Genes are ordered based on their phase in Constant Light (Vijayan et al., 2009), with the group of dusk and dawn genes indicated with colored bars next to the left heat map. Dusk genes increase in expression under Constant Light in wildtype cells and are maximally expressed when cells expect dusk (left panel, subjective dusk occurs at 36 hr in Constant Light). Note that these dynamics are very similar to the dynamics we observe in Low Light conditions in which the cells periodically experience 12 hr of darkness (Figure 2B, top panel). Dusk genes have constant low expression in OX-D53E cells without inducer which lack RpaA (center) and increase in expression when the RpaAP mimic RpaA-D53E is induced in this background (OX-D53E with inducer, right). (B) Data from (A) plotted for the representative dusk gene Synpcc7942_1567. (C) Data from (A) plotted for the representative dawn gene petA.
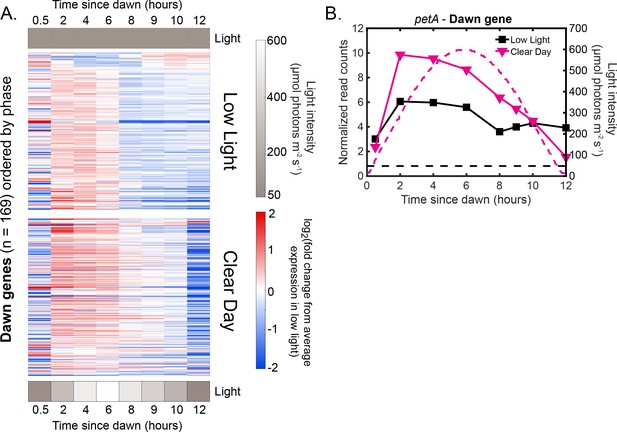
Dawn gene expression increases during the early part of Clear Day relative to Low Light conditions.
(A) Gene expression dynamics of dawn genes (=169) under Low Light (top) and Clear Day (bottom) conditions. Gene expression is quantified as the log fold change from the average expression of the gene over all time points in the Low Light condition (See Materials and methods - RNA sequencing for more details). Genes are plotted in the same order in both heat maps and were sorted by phase under Constant Light conditions (Vijayan et al., 2009). Most dawn genes are expressed more highly during the early part of Clear Day conditions compared to Low Light conditions. (B) Gene expression dynamics of the representative dawn gene petA under Low Light (black) and Clear Day (magenta) conditions as measured by RNA sequencing (left y-axis). The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis.
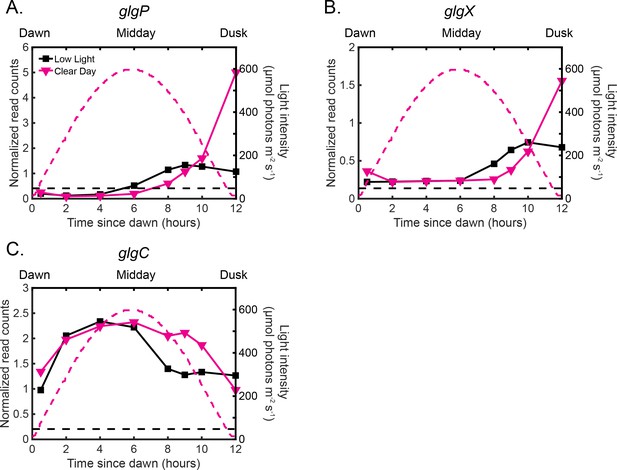
The gene expression dynamics of glycogen production and breakdown enzymes change in Clear Day conditions relative to Low Light conditions.
(A) Gene expression dynamics of the dusk gene glgP, encoding a key enzyme in glycogen breakdown, under Low Light (black) and Clear Day (magenta) conditions as measured by RNA sequencing (left y-axis). The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis. (B) Gene expression dynamics of the dusk gene glgX, a key enzyme in glycogen breakdown, measured and plotted as in (A). (C) Gene expression dynamics of the dusk gene glgC, a key enzyme in glycogen production, measured and plotted as in (A).
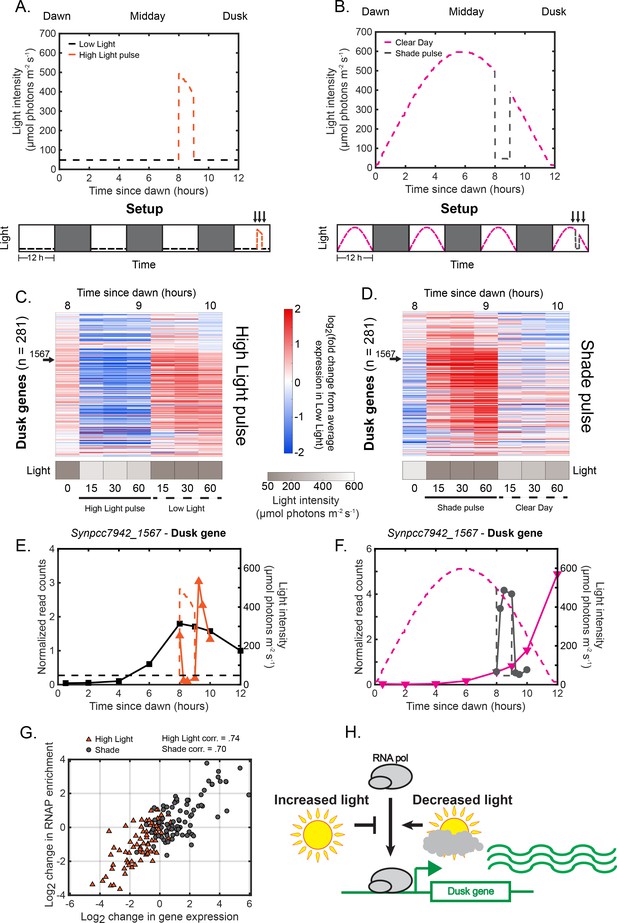
Rapid changes in light intensity modulate the recruitment of RNA polymerase to dusk genes to control dusk gene expression.
(A) Light intensity profiles of Low Light (black) and High Light pulse (orange) conditions, in units of mol photons m s (see Materials and methods - Calibrating light conditions for more details; light intensity values for pulse conditions available in Figure 3—source data 1). Experimental setup is displayed in the lower plot. Cells were grown for 12 hr under Low Light conditions (black dashed lines), followed by 12 hr of darkness (dark gray boxes) for three days, and then exposed to a High Light pulse (dashed orange lines) at 8 hr after dawn during the fourth light period for one hour before being returned to Low Light conditions. Cells were sampled immediately before, during, and after the High Light pulse (indicated by arrows above plot). (Caption continued on next page.). (B) Light intensity profiles of Clear Day (magenta) and Shade pulse (gray) conditions, in units of mol photons m s. Experimental setup is displayed in the lower plot. Cells were grown for 12 hr under Clear Day conditions (dashed magenta lines), followed by 12 hr of darkness (dark gray boxes) for three days, and then exposed to a Shade pulse (dashed gray lines) at 8 hr after dawn during the fourth light period for one hour before being returned to Low Light conditions. Cells were sampled immediately before, during, and after the High Light pulse (indicated by arrows above plot). (C) Gene expression dynamics of dusk genes (=281) under High Light pulse conditions. Gene expression is quantified as the log fold change from the average expression of the gene over all time points in the Low Light condition (see Materials and methods, RNA sequencing for more details; data available in Figure 3—source data 1). Light intensity at each time point in the High Light pulse condition is indicated in a grayscale heat map next to the corresponding time point. (D) Gene expression dynamics of dusk genes (=281) under Shade pulse conditions, plotted as in (C). Genes are ordered the same in (C) and (D), sorted by phase under Constant Light conditions (Vijayan et al., 2009). Data for the representative dusk gene Synpcc7942_1567 is indicated by arrows in (C) and (D). (E) Gene expression dynamics of the representative dusk gene Synpcc7942_1567 under Low Light (black) and High Light pulse (orange) conditions (left y-axis). The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis. (F) Gene expression dynamics of the representative dusk gene Synpcc7942_1567 under Clear Day (magenta) and Shade pulse (gray) conditions, plotted as in (E). (G) Correlation between change in dusk gene expression and the change in enrichment of RNAP upstream of that gene after rapid changes in light intensity. The change in gene expression of a dusk gene (x-axis) and the corresponding change in RNAP enrichment upstream of that gene (y-axis) from the original condition after 60 min in High Light (orange triangles) or Shade (gray circles), plotted for the 82 dusk genes with detectable RNAP peaks in their promoters. See Materials and methods, ChIP-seq analysis for more details. Data is available in Figure 3—source data 2. The correlation coefficient between change in RNAP enrichment and change in downstream gene expression for the High Light and Shade conditions is indicated above the plot. (H) Regulation of RNAP recruitment to dusk genes by changes in light intensity. Increases in light intensity tend to repress the recruitment of RNAP to dusk genes to repress dusk gene expression (High Light pulse, Clear Day - midday), while decreases in light intensity (Shade pulse, Sunset of the Clear Day) tend to promote the recruitment of RNAP to dusk genes to activate their expression.
-
Figure 3—source data 1
Normalized gene expression in High Light pulse and Shade pulse conditions.
- https://doi.org/10.7554/eLife.32032.013
-
Figure 3—source data 2
List of RNAP peaks, gene targets, and quantification of enrichment under High Light pulse and Shade pulse conditions.
- https://doi.org/10.7554/eLife.32032.014
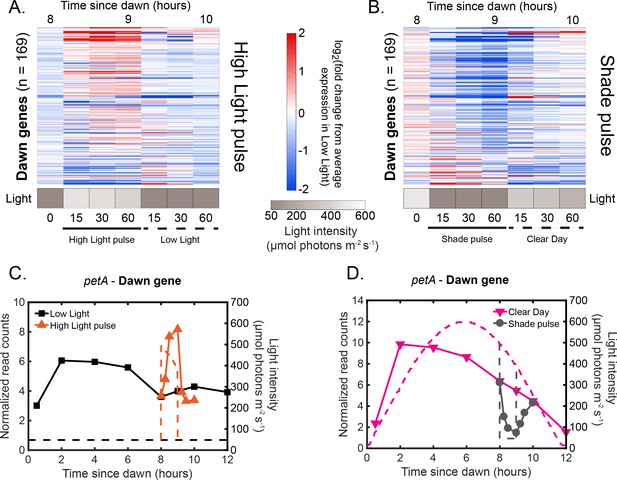
Rapid changes in light intensity affect dawn gene expression in an opposite direction compared to dusk gene expression.
(A) Gene expression dynamics of dawn genes (=169) under High Light pulse conditions. Gene expression is quantified as the log fold change from the average expression of the gene over all time points in the Low Light condition (See Materials and methods - RNA sequencing for more details). Light intensity at each time point in the High Light pulse condition is indicated in a grayscale heat map. Most dawn genes increase in expression after exposure to the High Light pulse. (B) Gene expression dynamics of all dawn genes (=169) under Shade pulse conditions, plotted as in (A). Genes are ordered the same in (A) and (B), sorted by phase under Constant Light conditions (Vijayan et al., 2009). Most dawn genes decrease in expression after exposure to the Shade pulse. (C) Gene expression dynamics of the representative dawn gene petA under Low Light (black) and High Light pulse (orange) conditions (left y-axis) as measured by RNA sequencing. The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis. (D) Gene expression dynamics of the representative dawn gene petA under Clear Day (magenta) and Shade pulse (gray) conditions, plotted as in (C).
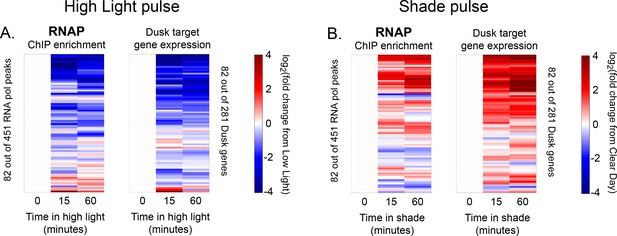
Changes in RNAP enrichment and downstream dusk gene expression after rapid changes in light intensity.
(A) Changes in enrichment of RNAP upstream of dusk genes during High Light pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map) for the 82 dusk genes with RNAP peaks in their promoters. ChIP enrichment (left heat map) is quantified as the log fold change of enrichment in High Light compared to enrichment at 8 hr since dawn in Low Light conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RNAP peak expressed as the log fold change of expression in High Light compared to expression at 8 hr since dawn in Low Light conditions (time zero). An RNAP peak and its target gene are aligned horizontally in the two heat maps so that they can be directly compared side-by-side. See Materials and methods, ChIP-seq analysis for more details. (B) Changes in enrichment of RNAP upstream of dusk genes during Shade pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map). ChIP enrichment (left heat map) is quantified as the log fold change of enrichment in Shade from enrichment at 8 hr since dawn in Clear Day conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RNAP peak expressed as the log fold change of expression in Shade from expression at 8 hr since dawn in Clear Day conditions (time zero). An RNAP peak and its target gene are aligned in the two heat maps. RNAP peaks and genes have the same order in (A) and (B). See Materials and methods, ChIP-seq analysis for more details. The correlation between RNAP enrichment change and downstream dusk gene expression reported in Figure 3G also holds after 15 min of exposure to High Light or Shade (High Light correlation = 0.64, Shade correlation = 0.68, changes after 15 min).
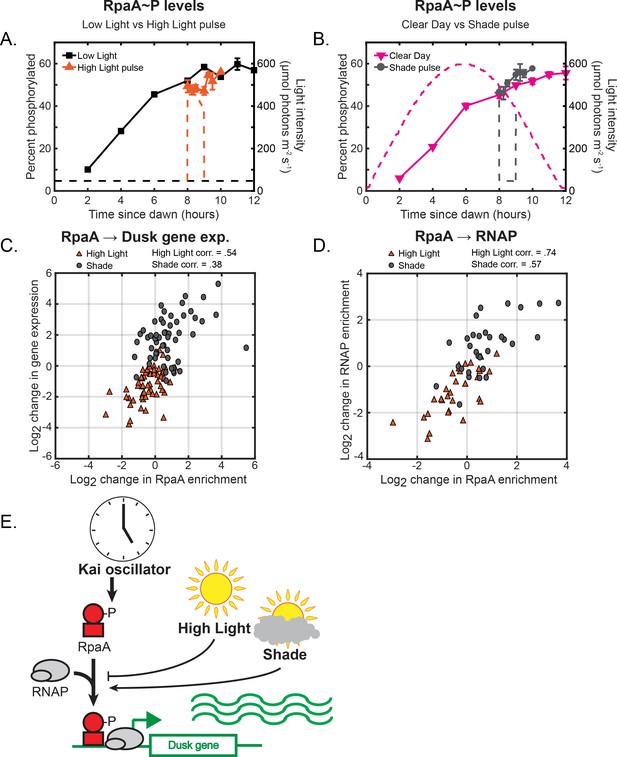
Changes in environmental light intensity regulate RpaAP DNA binding activity and RNAP recruitment to control dusk gene expression downstream of clock regulation of RpaA.
(A) Phosphorylation dynamics of RpaA under Low Light vs High Light pulse. Relative levels of phosphorylated RpaA were measured using Phos-tag Western blotting (left y-axis) in cells grown under Low Light conditions (black squares, see Figure 2A for Setup) or High Light pulse conditions (orange triangles, see Figure 3A for Setup). Each point represents the average of values measured in two independent Western blots, with error bars displaying the range of the measured values. See Materials and methods, Measurement of RpaAP and RpaBP levels for more details. Data is available in Figure 4—source data 1. The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis. (B) Phosphorylation dynamics of RpaA under Clear Day (magenta triangles, see Figure 2A for Setup) vs Shade pulse (gray circles, see Figure 3B for Setup) conditions, measured and plotted as in (A). (C) The change in enrichment of RpaA at a given peak upstream of a dusk gene (x-axis) and the corresponding change in expression of the downstream dusk gene (y-axis) from the original condition after 60 min in High Light (orange triangles) or Shade (gray circles), plotted for the 56 dusk genes with detectable RpaA peaks in their promoters. The correlation coefficient for the data taken in High Light and Shade conditions is indicated above the plot. See Materials and methods, ChIP-seq analysis for more details. Data is available in Figure 4—source data 2. (D) The change in enrichment of RpaA at a given peak upstream of a dusk gene (x-axis) and the corresponding change in RNAP enrichment upstream of the same gene (y-axis) from the original condition after 60 min in High Light (orange triangles) or Shade (gray circles), plotted for the 33 dusk genes with detectable RpaA and RNAP peaks in their promoters. The correlation coefficient for High Light and Shade data is indicated above the plot. See Materials and methods, ChIP-seq analysis for more details. (E) Model of regulation of dusk genes by RpaA under naturally-relevant conditions. The Kai PTO controls levels of RpaAP independent of changes in environmental light intensity. Changes in light intensity regulate the recruitment of RpaAP with RNAP to dusk genes to control their expression in response to environmental perturbations.
-
Figure 4—source data 1
Quantification of relative RpaA∼P levels.
- https://doi.org/10.7554/eLife.32032.020
-
Figure 4—source data 2
List of RpaA peaks, gene targets, and quantification of enrichment under High Light pulse and Shade pulse conditions.
- https://doi.org/10.7554/eLife.32032.021
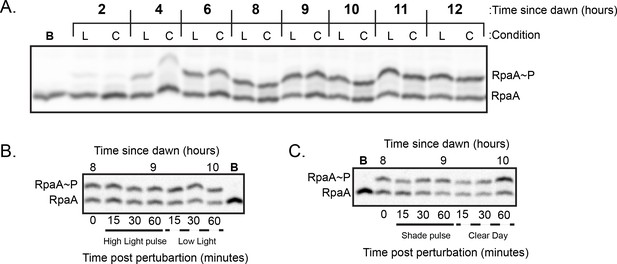
Representative Western blots used to quantify relative levels of RpaAP under dynamic light conditions.
(A) Representative Western Blot used to quantify levels of RpaAP under Low Light and Clear Day conditions. Lysates were prepared from cells harvested from either Low Light (L) or Clear Day (C) conditions at the indicated time and subject to Phos-tag electrophoresis and Western blotting with an anti-RpaA antibody (see Materials and methods). One sample was boiled prior to loading (Lane indicated with ‘B’) to identify the heat-labile band on the gel corresponding to RpaAP. (B) Representative Western Blot used to quantify levels of RpaAP under High Light pulse conditions. Time 0 refers to 8 hr since dawn under Low Light conditions. (C) Representative Western Blot used to quantify levels of RpaAP under Shade pulse conditions. Time 0 refers to 8 hr since dawn under Clear Day conditions.
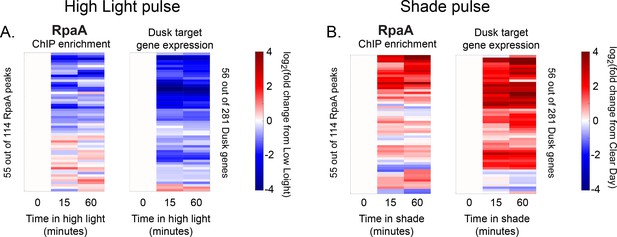
Changes in RpaA enrichment and downstream dusk gene expression after rapid changes in light intensity.
(A) Changes in enrichment of RpaA upstream of dusk genes during High Light pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map) for the 56 dusk genes with RpaA peaks in their promoters. ChIP enrichment (left heat map) is expressed as the log fold change of enrichment in High Light relative to enrichment at 8 hr since dawn in Low Light conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RpaA peak expressed as the log fold change of expression in High Light from expression at 8 hr since dawn in Low Light conditions (time zero). An RpaA peak and its target gene are aligned horizontally in the two heat maps. See Materials and methods, ChIP-seq analysis for more details. (B) Changes in enrichment of RpaA upstream of dusk genes during Shade pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map). ChIP enrichment (left heat map) is expressed as the log fold change of enrichment in Shade relative to enrichment at 8 hr since dawn in Clear Day conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RpaA peak expressed as the log fold change of expression in Shade from expression at 8 hr since dawn in Clear Day conditions (time zero). An RpaA peak and its target gene are aligned horizontally in the two heat maps. Peaks and genes have the same order in (A) and (B). See Materials and methods, ChIP-seq analysis for more details. The correlation between RpaA change and downstream dusk gene expression reported in Figure 4C also holds after 15 min of exposure to High Light or Shade (High Light correlation = 0.54, Shade correlation = 0.38, changes after 15 min).
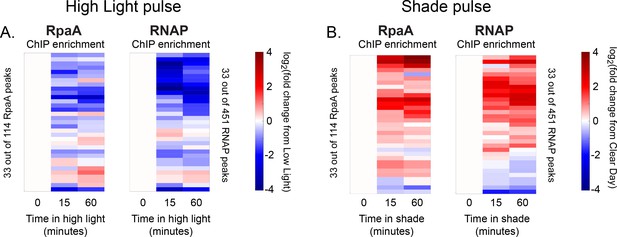
Changes in RpaA and RNA polymerase enrichment upstream of dusk genes after rapid changes in light intensity.
(A) Changes in enrichment of RpaA upstream of dusk genes during High Light pulse conditions (left heat map) and corresponding changes in RNAP enrichment upstream of the same gene (right heat map) for the 33 dusk genes with RpaA and RNAP peaks in their promoters. ChIP enrichment is quantified as the log fold change of enrichment in High Light from enrichment at 8 hr since dawn in Low Light conditions (time zero). RpaA and RNAP peaks upstream of the same dusk gene are aligned horizontally in the two heat maps. See Materials and methods, ChIP-seq analysis for more details. (B) Changes in enrichment of RpaA upstream of dusk genes during Shade pulse conditions (left heat map) and corresponding changes in RNAP enrichment upstream of the same gene (right heat map). ChIP enrichment is quantified as the log fold change of enrichment in Shade from enrichment at 8 hr since dawn in Clear Day conditions (time zero). RpaA and RNAP peaks upstream of the same dusk gene are aligned horizontally in the two heat maps. Peaks have the same order in (A) and (B). See Materials and methods, ChIP-seq analysis for more details. The correlation between RpaA and RNAP enrichment change upstream of dusk gene expressions reported in Figure 4D also holds after 15 min of exposure to High Light or Shade (High Light correlation = 0.56, Shade correlation = 0.42).
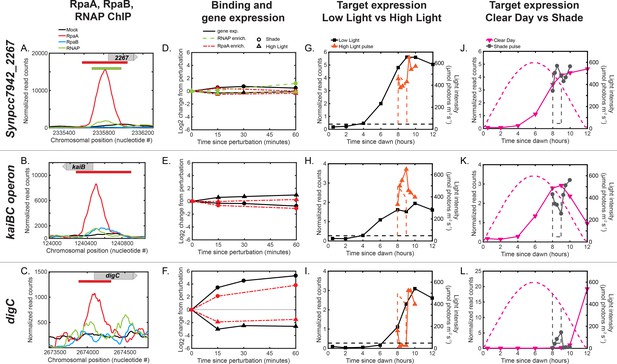
Multifactorial behavior of RpaAP at select promoters under changes in light intensity.
(A)-(C) Normalized ChIP-seq signal of RpaA (red), RpaB (blue), RNAP (green) and mock IP (black) upstream of the (A) the representative dusk gene Synpcc7942_2267, (B) the kaiBC operon and (C) another representative dusk gene, digC, at 8 hr since dawn in Low Light. The chromosomal position of the gene is located on the plot with a gray bar with an arrow indicating directionality of the gene. The location of RpaA and RNAP peaks are indicated on top of the plot with red (RpaA) and green (RNAP) bars. No RpaB peaks were found upstream of these genes. No RNAP peak was found upstream of kaiB or digC. See Materials and methods, ChIP-seq analysis for more details. (D)-(F) Changes in enrichment of RpaA (red) and RNAP (green) and downstream gene expression (black) after exposure to the High Light pulse (triangles) or the Shade pulse (circles) for (D) Synpcc7942_2267, (E) the kaiBC operon, and (F) digC. See Materials and methods, ChIP-seq analysis for more details. (G)-(L) Gene expression dynamics of Synpcc7942_2267 (G,J) kaiB (H,K) and digC (I,L) under Low Light vs High Light pulse (G–I) and Clear Day vs Shade pulse (J–L) conditions. RpaA binding does not change upstream of the dusk genes Synpcc7942_2267 and kaiB after changes in light (D,E), and the expression of these genes does not change significantly in response to changes in light intensity (G,J;H,K). In contrast, RpaA binding changes significantly (F) upstream of the light-responsive dusk gene digC (I,L).
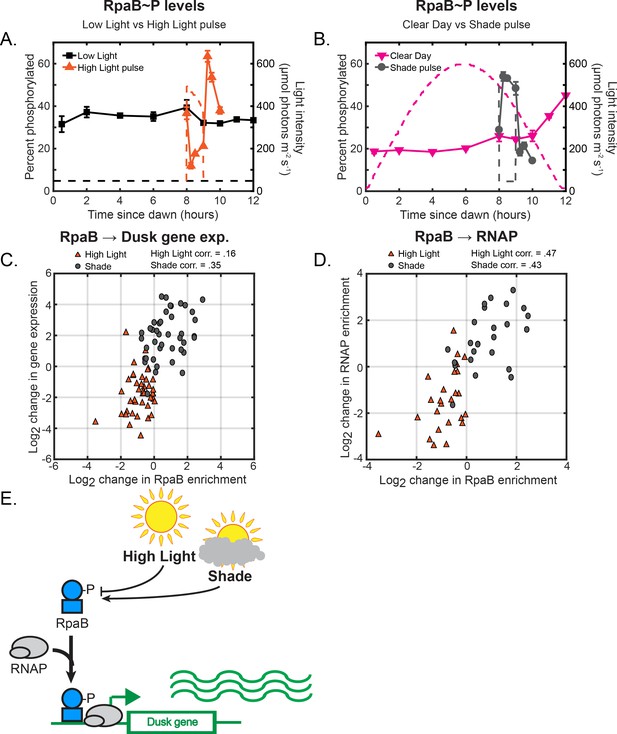
Light-induced changes in RpaBP levels modulate RpaB and RNAP binding upstream of dusk genes to directly regulate dusk gene expression in response to light.
(A) Phosphorylation dynamics of RpaB under Low Light vs High Light pulse. Relative levels of phosphorylated RpaB were measured using Phos-tag Western blotting (left y-axis) in cells grown under Low Light conditions (black squares, see Figure 2A for Setup) or High Light pulse conditions (orange triangles, see Figure 3A for Setup). Each point represents the average of values measured in two independent Western blots, with error bars displaying the range of the measured values. See Materials and methods, Measurement of RpaAP and RpaBP levels for more details. Data is available in Figure 5—source data 1. The light profile for each condition is plotted as dashed lines of the same color with values corresponding to the right y-axis. (B) Phosphorylation dynamics of RpaB under Clear Day (magenta triangles, see Figure 2A for Setup) vs Shade pulse (gray circles, see Figure 3B for Setup) conditions, measured and plotted as in (A). (C) The change in enrichment of RpaB at a given peak upstream of a dusk gene (x-axis) and the corresponding change expression of the downstream dusk gene (y-axis) from the original condition after 60 min in High Light (orange triangles) or Shade (gray circles), plotted for the 42 dusk genes with detectable RpaB peaks in their promoters. The correlation coefficient for High Light and Shade data is indicated above the plot. See Materials and methods, ChIP-seq analysis for more details. Data is available in Figure 5—source data 2. (D) The change in enrichment of an RpaB at a given peak upstream of a dusk gene (x-axis) and the corresponding change in RNAP enrichment upstream of the same gene (y-axis) from the original condition after 60 min in High Light (orange triangles) or Shade (gray circles), plotted for the 27 dusk genes with detectable RpaB and RNAP peaks in their promoters. he correlation coefficient for High Light and Shade data is indicated above the plot. See Materials and methods, ChIP-seq analysis for more details. (E) Model of regulation of dusk genes by RpaB under naturally-relevant conditions. Changes in light regulate RpaBP levels. RpaBP binds with RNAP to dusk genes to control their expression in response to environmental perturbations.
-
Figure 5—source data 1
Quantification of relative RpaB∼P levels.
- https://doi.org/10.7554/eLife.32032.026
-
Figure 5—source data 2
List of RpaB peaks, gene targets, and quantification of enrichment under High Light pulse and Shade pulse conditions.
- https://doi.org/10.7554/eLife.32032.027
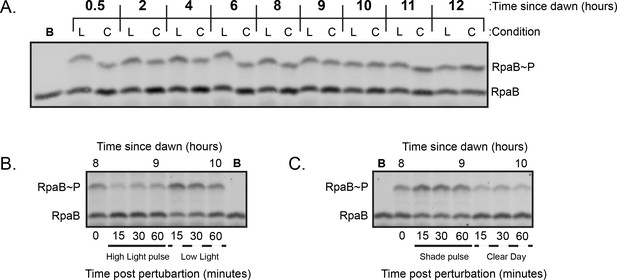
Representative Western blots used to quantify relative levels of RpaBP under dynamic light conditions.
(A) Representative Western Blot used to quantify levels of RpaBP under Low Light and Clear Day conditions. Lysates were prepared from cells harvested from either Low Light (L) or Clear Day (C) conditions at the indicated time and subject to Phos-tag electrophoresis and Western blotting with an anti-RpaB antibody (see Methods). One sample was boiled prior to loading (Lane indicated with ’B’) to identify the heat-labile band on the gel corresponding to RpaBP. (B) Representative Western Blot used to quantify levels of RpaBP under High Light pulse conditions. Time 0 refers to 8 hr since dawn under Low Light conditions. (C) Representative Western Blot used to quantify levels of RpaBP under Shade pulse conditions. Time 0 refers to 8 hr since dawn under Clear Day conditions.
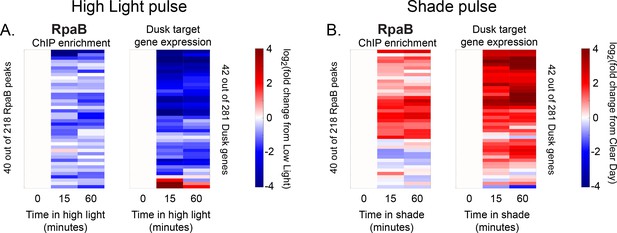
Changes in RpaB enrichment and downstream dusk gene expression after rapid changes in light intensity.
(A) Changes in enrichment of RpaB upstream of dusk genes during High Light pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map) for the 42 dusk genes with RpaB peaks in their promoters. ChIP enrichment (left heat map) is quantified as the log fold change of enrichment in High Light from enrichment at 8 hr since dawn in Low Light conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RpaB peak quantified as the log fold change of expression in High Light from expression at 8 hr since dawn in Low Light conditions (time zero). An RpaB peak and its target gene are aligned horizontally in the two heat maps. See Materials and methods, ChIP-seq analysis for more details. (B) Changes in enrichment of RpaB upstream of dusk genes during Shade pulse conditions (left heat map) and corresponding changes in target dusk gene expression (right heat map). ChIP enrichment (left heat map) is quantified as the log fold change of enrichment in Shade from enrichment at 8 hr since dawn in Clear Day conditions (time zero). The right heat map shows the change in expression of the gene target of the corresponding RpaB peak quantified as the log fold change of expression in Shade from expression at 8 hr since dawn in Clear Day conditions (time zero). An RpaB peak and its target gene are aligned horizontally in the two heat maps. Peaks and genes have the same order in (A) and (B). See Materials and methods, ChIP-seq analysis for more details. The correlation between RpaB change and downstream dusk gene expression reported in Figure 5C also holds after 15 min of exposure to High Light or Shade (High Light correlation = 0.16, Shade correlation = 0.35, changes after 15 min).
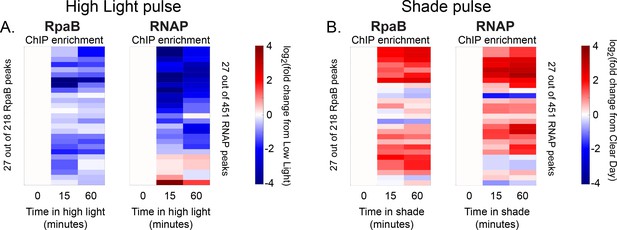
Changes in RpaB and RNA polymerase enrichment upstream of dusk genes after rapid changes in light intensity.
(A) Changes in enrichment of RpaB upstream of dusk genes during High Light pulse conditions (left heat map) and corresponding changes in RNAP enrichment upstream of the same gene (right heat map) for the 27 dusk genes with RpaB and RNAP peaks in their promoters. ChIP enrichment is expressed as the log fold change of enrichment in High Light from enrichment at 8 hr since dawn in Low Light conditions (time zero). RpaB and RNAP peaks upstream of the same dusk gene are aligned horizontally in the two heat maps. See Materials and methods, ChIP-seq analysis for more details. (B) Changes in enrichment of RpaB upstream of dusk genes during Shade pulse conditions (left heat map) and corresponding changes in RNAP enrichment upstream of the same gene (right heat map). ChIP enrichment is expressed as the log fold change of enrichment in Shade from enrichment at 8 hr since dawn in Clear Day conditions (time zero). RpaB RNAP peaks upstream of the same dusk gene are aligned horizontally in the two heat maps. Peaks have the same order in (A) and (B). See Materials and methods, ChIP-seq analysis for more details. The correlation between RpaA and RNAP enrichment change upstream of dusk gene expressions reported in Figure 5D also holds after 15 min of exposure to High Light or Shade (High Light correlation = 0.22, Shade correlation = 0.42).
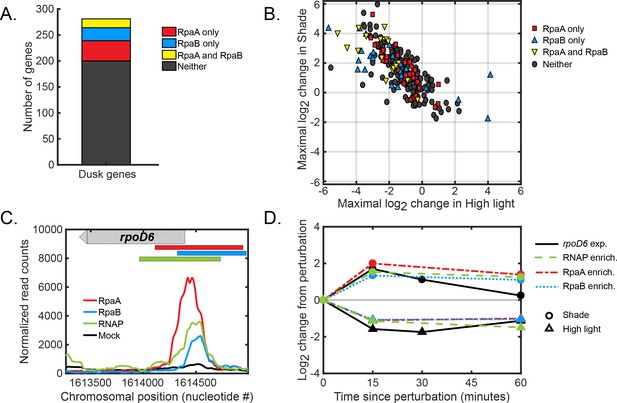
Global regulation of dusk gene expression in response to light changes.
(A) Number of dusk gene targets of RpaA only (red), RpaB only (blue), RpaA and RpaB (yellow), or neither (black). Target genes of binding sites of RpaA and RpaB were determined using chromatin immunoprecipitation followed by sequencing under several different light conditions (see Materials and methods, ChIP-seq analysis, for more details. See Figure 4—source data 2 or Figure 5—source data 2 for full lists of RpaA and RpaB peaks associated with dusk genes). (B) Light-responsive changes in gene expression of dusk genes. For each dusk gene, we calculated the maximal log change in expression during the High Light pulse (x-axis) or Shade pulse (y-axis) from 8 hr since dawn in the Low light or Clear day conditions, respectively, using the data from Figure 3. (C) Normalized ChIP-seq signal of RpaA (red), RpaB (blue), RNAP (green) and mock IP (black) upstream of the dusk sigma factor gene rpoD6 at 8 hr since dawn in Low Light. The chromosomal location of the gene is located on the plot with a gray bar with an arrow indicating directionality of the gene. The location of RpaA, RpaB, and RNAP peaks are indicated on top of the plot with red (RpaA), blue (RpaB), and green (RNAP) bars. See Materials and methods, ChIP-seq analysis for more details. (D) Changes in enrichment upstream of rpoD6 of RpaA (red), RpaB (blue), and RNAP (green) and changes in rpoD6 gene expression (black) after exposure to the High Light pulse (triangles) or the Shade pulse (circles). See Materials and methods, ChIP-seq analysis for more details.
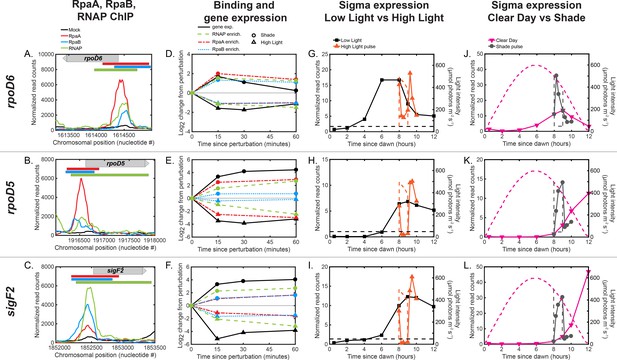
Regulation of dusk sigma factor gene expression by RpaA and RpaB.
(A)-(C) Normalized ChIP-seq signal of RpaA (red), RpaB (blue), RNAP (green) and mock IP (black) upstream of the sigma factor genes (A) rpoD6, (B) rpoD5, and (C) sigF2. The location of the gene is located on the plot with a gray bar with an arrow indicating directionality of the gene. The location of RpaA, RpaB, and RNAP peaks are indicated on top of the plot with red (RpaA), blue (RpaB), and green (RNAP) bars. See Materials and methods, ChIP-seq analysis for more details. (D)-(F) Changes in enrichment of RpaA (red), RpaB (blue), and RNAP (green) and downstream sigma factor gene expression (black) after exposure to the High Light pulse (triangles) or the Shade pulse (circles) upstream of rpoD6 (D), rpoD5 (E), and sigF2 (F). See Materials and methods, ChIP-seq analysis for more details. (G)-(L) Gene expression dynamics of rpoD6 (G,J), rpoD5 (H,K), and sigF2 (I,L) under Low Light vs High Light pulse (G)-(I) and Clear Day vs Shade pulse (J)-(L) conditions. RpaA and RpaB binding changes in a correlated manner upstream of these genes. RpaA and RpaB binding also correlates with changes in RNAP enrichment and sigma factor expression levels.
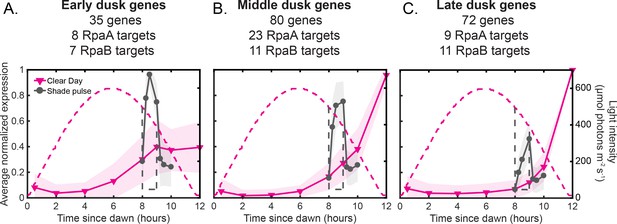
Dusk genes group into three major clusters that show distinct and coordinated responses to changes in light intensity.
(A) Average expression profiles of genes belonging to the Early dusk gene cluster under Clear Day (magenta) and Shade pulse (gray) conditions (left y-axis). Dusk genes were grouped using k-means clustering of their normalized expression dynamics in response to the four light conditions of this study and perturbations of RpaA activity in Constant Light conditions (Figure 7—figure supplement 1, [Markson et al., 2013]), and clusters were named based on their order of activation. See Materials and methods - K-means clustering for more details, and Figure 7—source data 1 for full lists of genes in each cluster. The number of genes within the cluster, as well as the number of genes with an RpaA or RpaB peak in their promoters (targets) is listed. The expression values of each gene across all four light conditions in this work were normalized to a range of 0 to 1, and the normalized expression values were averaged within each cluster (solid lines). The shaded region on the plot indicates the standard deviation of the normalized expression values within the cluster. The light intensity profile for each condition is plotted as dashed lines in the same color with values corresponding to the right y-axis. (B) Average expression profiles of genes belonging to the Middle dusk gene cluster under Clear Day (magenta) and Shade pulse (gray) conditions (left y-axis), presented as in (A). (C) Average expression profiles of genes belonging to the Late dusk gene cluster under Clear Day (magenta) and Shade pulse (gray) conditions (left y-axis), presented as in (A).
-
Figure 7—source data 1
Lists of genes belonging to the Early, Middle, and Late dusk clusters, and scaled gene expression values.
- https://doi.org/10.7554/eLife.32032.032
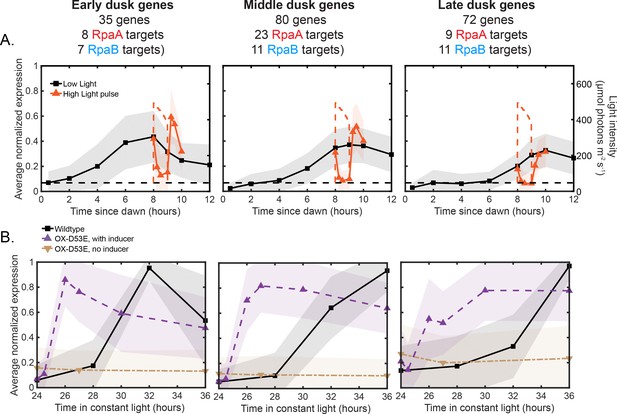
Average expression profiles of the major dusk gene clusters under various conditions.
(A) Average expression profiles of the Early (left plot), Middle (middle plot), and Late (right plot) dusk gene clusters under Low Light (black) and High Light pulse (orange) conditions (left y-axis). The expression values of each gene across all four light conditions in this work were normalized to a range of 0 to 1, and the normalized expression values were averaged within each cluster. The shaded region of the plot indicates the standard deviation of the normalized expression values within the cluster. Lists of genes belonging to each cluster and the scaled expression values are available in Figure 7—source data 1. The light intensity profile for each condition is plotted as dashed lines in the same color with values corresponding to the right y-axis. (B) Average expression profiles of the Early (left plot), Middle (middle plot), and Late (right plot) dusk gene clusters in Constant Light conditions in wildtype and OX-D53E cells (rpaA-, kaiBC-, Ptrc::rpaA(D53E)) (data from [Markson et al., 2013]). The OX-D53E strain allows experimental control of RpaA activity via IPTG-inducible expression of the RpaA phosphomimetic RpaA-D53E in cells that lack wildtype RpaA. Plotted are average cluster expression in wildtype cells in Constant Light conditions (black squares), OX-D53E cells without inducer (RpaA phosphomimetic not induced, brown downward triangles), and OX-D53E cells with inducer (RpaA phosphomimetic induced, purple upward triangles). The expression values of each gene within each strain in Constant Light (wildtype or OX-D53E) were separately normalized to a range of 0 to 1, and the normalized expression values were averaged within each cluster. Lists of genes belonging to each cluster and the scaled expression values are available in Figure 7—source data 1. The shaded region on the plot indicates the standard deviation of the normalized expression values within the cluster.
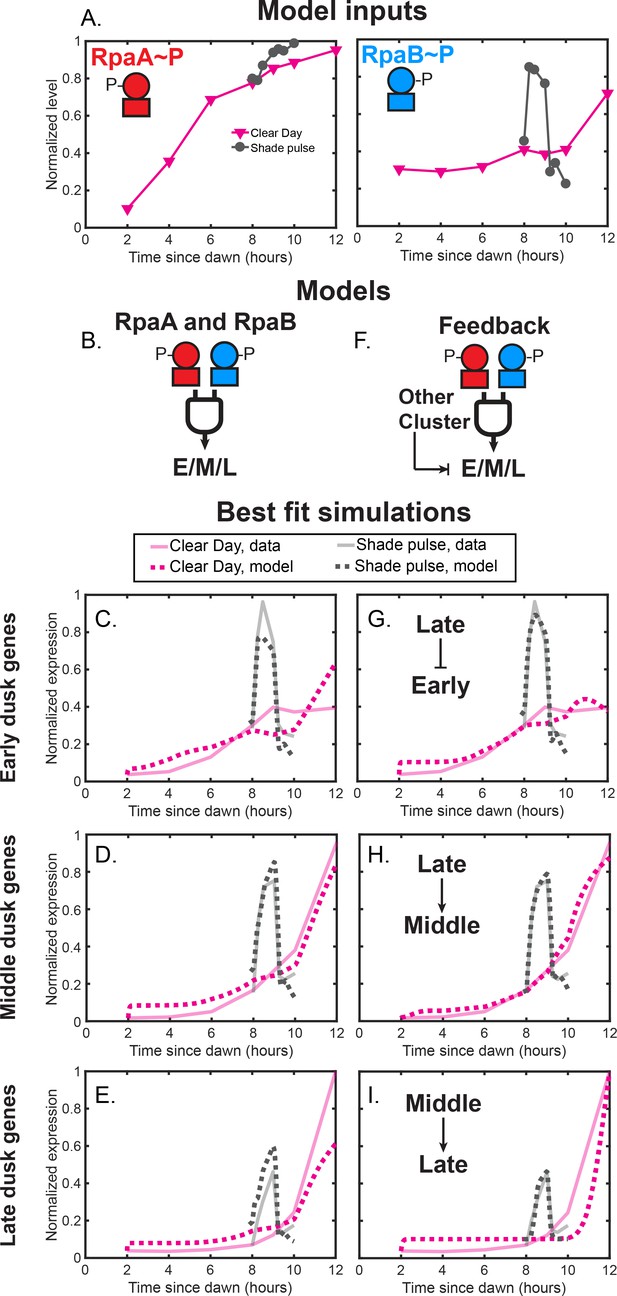
Phenomenological modeling of the activation of clusters of light-responsive dusk genes.
(A) Normalized RpaAP levels (left plot) and RpaBP levels (right plot) under Clear Day (magenta) and Shade pulse (gray) conditions used as input for mathematical models of dusk gene expression. RpaAP or RpaBP levels from all four light conditions were normalized to a range of 0 to 1. (B) In the ‘RpaA and RpaB’ models, RpaAP and RpaBP jointly activate the expression of the Early (E), Middle (M), or Late (L) cluster. See Materials and methods - Mathematical modeling for more details. (C) Simulations of the best fit ‘RpaA and RpaB’ model for the Early dusk genes. Average cluster expression data is shown as faded solid lines, and the best fit simulations are shown as dotted lines. Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. See Materials and methods - Mathematical modeling for more details. (D) Simulations of the best fit ‘RpaA and RpaB’ model for the Middle dusk genes, plotted as in (C). (E) Simulations of the best fit ‘RpaA and RpaB’ model for the Late dusk genes, plotted as in (C). (F) In the ‘Feedback’ models, another cluster activates or represses the expression of the Early (E), Middle (M), or Late (L) cluster alongside joint activation by RpaAP and RpaBP. (G) Simulations of the best fit ‘Feedback’ model for the Early dusk genes, plotted as in (C). In this model, Late cluster expression represses Early cluster expression alongside activation by RpaAP and RpaBP. (H) Simulations of the best fit ‘Feedback’ model for the Middle dusk genes, plotted as in (C). In this model, Late cluster expression activates Middle cluster expression alongside activation by RpaAP and RpaBP. (I) Simulations of the best fit ‘Feedback’ model for the Late dusk genes, plotted as in (C). In this model, Middle cluster expression activates Late cluster expression alongside activation by RpaAP and RpaBP.
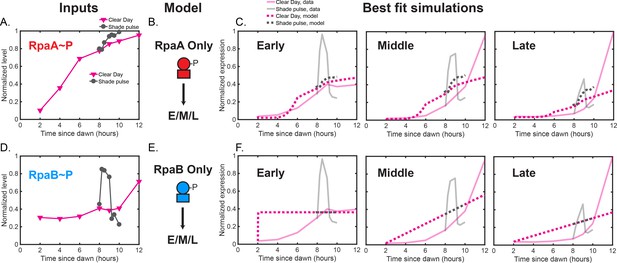
Best fit simulations of ‘RpaA-only’ and ‘RpaB-only’ models in which RpaAP or RpaBP solely activates the expression of the dusk gene clusters.
(A) Normalized RpaAP levels under Clear Day (magenta) and Shade pulse (gray) conditions used as input for mathematical models of dusk gene expression. RpaAP levels from all four light conditions were normalized to a range of 0 to 1. (B) In the ‘RpaA-only’ models, RpaAP activates the expression of the Early (E), Middle (M), or Late (L) cluster. (C) Simulations (dotted lines) of best fit RpaA-only models for Clear Day and Shade pulse data (solid lines) for the Early (Left plot), Middle (middle plot), and Late (right plot) dusk genes. Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (D) Normalized RpaBP levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input. (E) Model schematic. Dusk gene expression under Clear Day and Shade pulse conditions was modeled as an activation Hill function of RpaBP levels only. (F) Simulations (dotted lines) of best fit RpaB-only models for Clear Day and Shade pulse data (solid lines) for the Early (Left plot), Middle (middle plot), and Late (right plot) dusk genes. Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray.
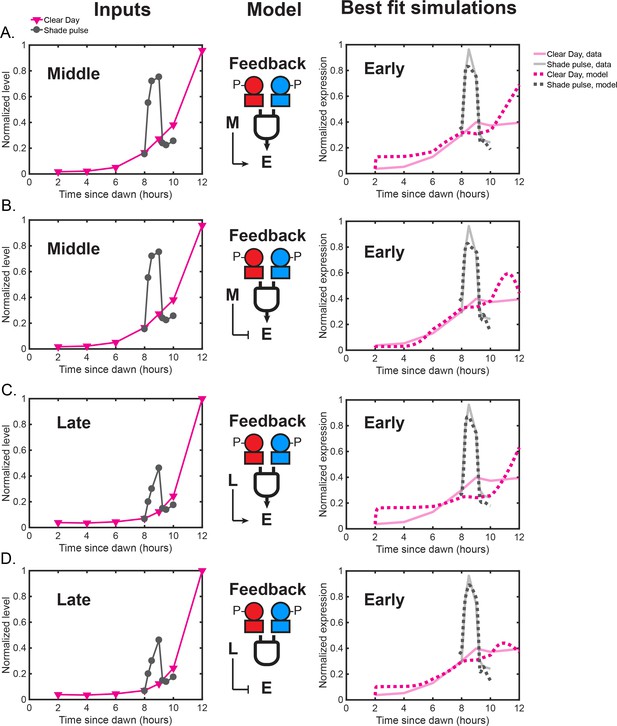
Models in which either the Middle or Late cluster feeds back to influence Early cluster expression.
(A) Feedback model in which the expression of the Early dusk cluster is an activation Hill function of Middle gene expression and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Middle cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input in addition to the RpaAP and RpaBP dynamics shown in Figure 8A of the main text. The right plot shows the average expression values of the Early cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (B) Feedback model in which Early gene cluster expression is a repression Hill function of Middle cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (A). (C) Feedback model in which Early cluster expression is an activation Hill function of Late cluster expression levels and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Late cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input. The right plot shows the average expression values of the Early cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (D) Feedback model in which Early cluster expression is a repression Hill function of Late cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (C). A model with an incoherent feedforward architecture in which the Late cluster represses Early cluster expression (D) best recapitulates the difference of Early cluster responses to Shade and Clear Day Sunset. During the Shade pulse, the Late cluster levels do not reach high enough levels to inhibit Early cluster expression, but at Sunset in Clear Day, Late cluster levels reach high enough levels to repress the expression of the Early cluster.
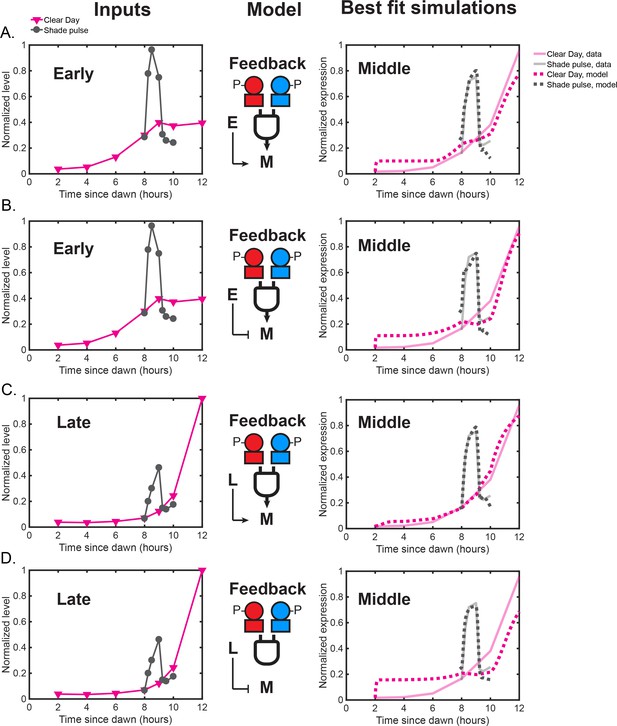
Models in which either the Early or Late cluster feeds back to influence Middle cluster expression.
(A) Feedback model in which the expression of the Middle dusk cluster is an activation Hill function of Early gene expression and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Early cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input in addition to the RpaAP and RpaBP dynamics shown in Figure 8A of the main text. The right plot shows the average expression values of the Middle cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (B) Feedback model in which Middle gene cluster expression is a repression Hill function of Early cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (A). (C) Feedback model in which Middle cluster expression is an activation Hill function of Late cluster expression levels and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Late cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input. The right plot shows the average expression values of the Middle cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (D) Feedback model in which Middle cluster expression is a repression Hill function of Late cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (C).
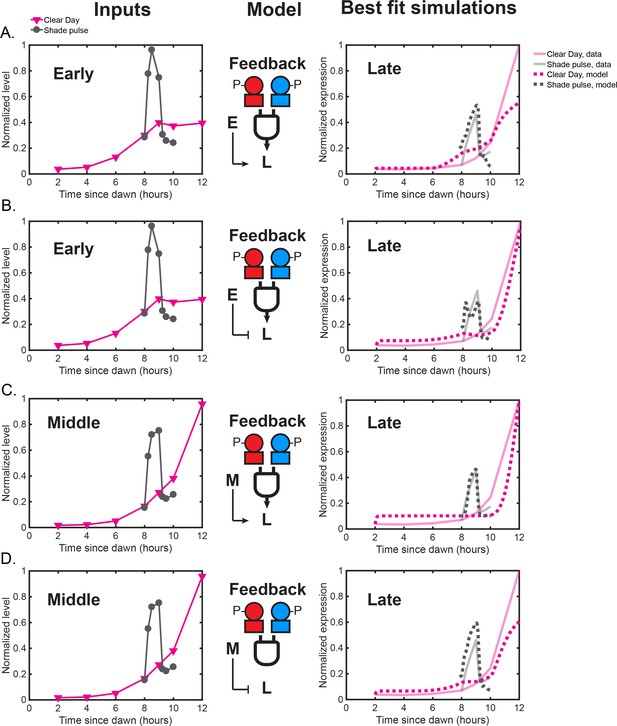
Models in which either the Early or Middle cluster feeds back to influence Late cluster expression.
(A) Feedback model in which the expression of the Late dusk cluster is an activation Hill function of Early gene expression and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Early cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input in addition to the RpaAP and RpaBP dynamics shown in Figure 8A of the main text. The right plot shows the average expression values of the Middle cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (B) Feedback model in which Late gene cluster expression is a repression Hill function of Early cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (A). (C) Feedback model in which Late cluster expression is an activation Hill function of Middle cluster expression levels and an activation Hill function of both RpaAP and RpaBP. The left plot shows normalized Middle cluster expression levels under Clear Day (magenta) and Shade pulse (gray) conditions used as model input. The right plot shows the average expression values of the Late cluster data (solid transparent lines), and the simulation produced the best fit model (dotted lines). Data for Clear Day conditions are plotted in magenta, and Shade pulse in gray. (D) Feedback model in which Late cluster expression is a repression Hill function of Middle cluster expression levels and an activation Hill function of both RpaAP and RpaBP, presented as in (C). A model with an coherent feedforward architecture where the Middle cluster activates the Late cluster (C) best recapitulates the difference of the Late cluster responses to Shade and Clear Day Sunset. During the Shade pulse, the Middle cluster levels do not reach high enough levels to allow for strong activation of Late cluster expression, but at Sunset in Clear Day, Middle cluster levels reach high enough levels to activate the expression of the Late cluster.
Tables
| Reagent type or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Synechococcus elongatus) | PCC 7942 (wild-type) | ATCC | Cat. Num. 33912 | |
| Strain, strain background (Escherichia coli) | Tuner (DE3) | EMD Millipore | Cat. Num. 70263 | |
| Gene (S. elongatus) | RNA polymerase Beta’ subunit | N/A | Cyanobase: Synpcc7942_1524 | |
| Gene (S. elongatus) | rpaB | N/A | Cyanobase: Synpcc7942_1453 | |
| Recombinant DNA reagent | RNA polymerase beta prime subunit FLAG | This paper | Addgene: 102337 | Plasmid encoding C-terminal FLAG tag RNA polymerase Beta’ subunit (Synpcc7942_1524) with Kan selection marker, targeted to integrate at native gene locus |
| Recombinant DNA reagent | pET-48b(+) | EMD Millipore | Cat. Num. 71462 | |
| Renetic reagent (S. elongatus) | EOC 398 and EOC 399 | This paper | S. elongatus PCC7942 transformed with RNA polymerase beta prime subunit FLAG plasmid. Confirmed by PCR and Western blot. | |
| Antibody | anti-RpaB | This paper | Anti-RpaB serum was produced by Cocalico Biologicals. Anti-RpaB was affinity purified as described in this work. | |
| Antibody | anti-RpaA | This paper | Anti-RpaA serum was produced by Cocalico Biologicals as described in Markson et al., 2013. Anti-RpaA was affinity purified as described in this work. | |
| Antibody | FLAG M2 mouse monoclonal antibody | Sigma Aldrich | Cat. Num. F3165 | |
| Software, algorithm | Imagequant | GE Healthcare | ||
| Software, algorithm | Bowtie | PMID: 19261174 | ||
| Software, algorithm | Peak-Seq | PMID: 19122651 | ||
| Software, algorithm | MATLAB | MathWorks | ||
| Commercial assay or kit | RNeasy Mini kit | Qiagen | Cat. Num. 74104 | |
| Commercial assay or kit | Ribo-Zero bacteria rRNA removal kit | Illumina | Cat. Num. MRZMB126 | |
| Commercial assay or kit | Truseq Stranded mRNA sample prep kit | Illumina | Cat. Num. 20020594 | |
| Commercial assay or kit | NEBNext Ultra II DNA library prep kit | New England Biolabs | Cat. Num. E7645S | |
| Chemical compound, drug | Phos-tagAcrylamide AAL-107 | Wako Pure Chemical Industries | Cat. Num. 304–93521 |
Fitting bounds.
Bounds used for fitting the variables in our simple model of gene expression. H is the Hill coefficient, is the max transcription rate, is the decay/dilution rate, is the background transcription rate, and is a coefficient of activation/repression (see equations 1-3, p. 1–3). The units of , , and are normalized expression/hr; is in normalized expression units.
| Variable | Lower bound | Upper bound |
|---|---|---|
| H | 0 | 7 |
| 0 | 80 | |
| 0 | 80 | |
| 0 | 10 | |
| 0 | 1 |
Fitting results.
The definitions of the variables are given in Equations 1-3, p. 1–3. The error is defined as the square root of the sum of the squared deviations between simulation and data.
| Model | Cluster | Figure | Error | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RpaA-only | Early | 7D | 0.71 | 37.54 | 72.71 | 0.71 | 6.76 | - | - | - | - | 0.85 |
| RpaB-only | Early | 7-Fig. Supp. 2C | 0.37 | 24.03 | 78.62 | - | - | 0.37 | 0.78 | - | - | 1.01 |
| RpaA and RpaB | Early | 7G | 0.35 | 51.28 | 37.76 | 0.35 | 4.19 | 0.8 | 2.5 | - | - | 0.41 |
| Feedback, M act. | Early | 7-Fig. Supp. 3A | 0.01 | 55.85 | 30.01 | 0.01 | 0.3 | 0.87 | 2.38 | 0.06 | 2.47 | 0.37 |
| Feedback, M rep. | Early | 7-Fig. Supp. 3B | 0.67 | 58.69 | 38.89 | 0.67 | 6.96 | 0.62 | 2.47 | 0.96 | 7 | 0.24 |
| Feedback, L act. | Early | 7-Fig. Supp. 3C | 0.2 | 35.87 | 19.03 | 0.2 | 4.43 | 0.98 | 3.35 | 0.05 | 6.15 | 0.38 |
| Feedback, L rep. | Early | 7I, 7-Fig. Supp. 3D | 0.75 | 69.34 | 42.68 | 0.75 | 6.22 | 0.59 | 3.53 | 0.71 | 2.39 | 0.21 |
| RpaA-only | Middle | 7D | 0.79 | 37.95 | 63 | 0.79 | 6.76 | - | - | - | - | 0.86 |
| RpaB-only | Middle | 7-Fig. Supp. 2C | 0.26 | 0.03 | - | - | - | 0.26 | 5.6 | - | - | 0.85 |
| RpaA and RpaB | Middle | 7G | 1 | 57.46 | 25.97 | 1 | 4.96 | 0.52 | 4.12 | - | - | 0.29 |
| Feedback, E act. | Middle | 7-Fig. Supp. 4A | 0.8 | 23.73 | 22.19 | 0.8 | 6.96 | 0.49 | 4.53 | 0.21 | 6.35 | 0.32 |
| Feedback, E rep. | Middle | 7-Fig. Supp. 4B | 0.73 | 71.08 | 39.24 | 0.73 | 5.14 | 0.53 | 6.58 | 0.74 | 0.88 | 0.35 |
| Feedback, L act. | Middle | 7I, 7-Fig. Supp. 4C | 0.18 | 78.63 | 76.5 | 0.18 | 6.09 | 0.33 | 2.64 | 0.16 | 1.55 | 0.16 |
| Feedback, L rep. | Middle | 7-Fig. Supp. 4D | 0.68 | 31.02 | 17.98 | 0.68 | 3.34 | 0.57 | 6.79 | 1 | 0 | 0.44 |
| RpaA-only | Late | 7D | 0.96 | 39.82 | 64.37 | 0.96 | 6.7 | - | - | - | - | 0.78 |
| RpaB-only | Late | 7-Fig. Supp. 2C | 0.05 | 0 | 0 | - | - | 0.05 | 0.68 | - | - | 0.79 |
| RpaA and RpaB | Late | 7G | 0.95 | 77.65 | 67.1 | 0.95 | 7 | 0.48 | 5.9 | - | - | 0.5 |
| Feedback, E act. | Late | 7-Fig. Supp. 5A | 0.99 | 23.93 | 20.01 | 0.99 | 5.8 | 0.4 | 6.95 | 0.18 | 6.77 | 0.53 |
| Feedback, E rep. | Late | 7-Fig. Supp. 5B | 0.76 | 59.81 | 18.43 | 0.76 | 6.22 | 0.69 | 6.13 | 0.47 | 3.12 | 0.29 |
| Feedback, M act. | Late | 7I, 7-Fig. Supp. 5C | 0.37 | 27.3 | 16.09 | 0.37 | 3.72 | 0.01 | 3.46 | 0.91 | 6.23 | 0.22 |
| Feedback, M rep. | Late | 7-Fig. Supp. 5D | 0.86 | 25.1 | 14.46 | 0.86 | 6.92 | 0.48 | 7 | 1 | 0 | 0.52 |
Parts for controllable light source.
The table includes the parts chosen for their specific properties. The remaining parts, such as wires, heat shrink tubing, thermal paste for mounting the LEDs on the heat sinks, proto-boards, and housing are quite general and specific brands are unnecessary.
| Part name | Digikey part number | Current price ($) | Quantity |
|---|---|---|---|
| PWR SUP MEDICAL 18V 8.3A 150W | EPS439-ND | 73.71 | 1 |
| CONN RCPT 8CONT DIN SLD PNL MNT | SC2007-ND | 5.64 | 1 |
| LEDDynamics Flexblock BUCK BOOST 48V, 700 mA | 788–1038-ND | 19.99 | 4 |
| AD7376 digital potentiometer | AD7376ARWZ10-ND | 8.66 | 4 |
| AC to DC power supply, 10VDC, 275 mA | 993–1233-ND | 4.68 | 2 |
| BXRA-30E1200-B-03, Bridgelux, Warm white, LED | Not sold at Digikey. | ||
| Need to order from: | 10.47 | 4 | |
| AMBIT ELECTRONICS, INC. | |||
| Aavid thermalloy Spotlight 47W heat sink | 1061–1092-ND | 9.50 | 4 |
| Arduino Uno Board Rev3 | 1050–1024-ND | 21.49 | 1 |
Wiring the FlexBlock LED driver.
The FlexBlock LED driver needs to be connected in a ’boost only’ configuration (see spec sheet for more details), with connections as shown.
| Line | Connection |
|---|---|
| DIM GND | GND of 10 V power supply/Arduino |
| DIM | Wipe of AD7376 potentiometer (Pin 16) |
| Vin+ | +of 18V power supply AND + of LED array |
| Vin- | GND of 18V power supply |
| LED+ | NC (not connected) |
| LED- | - of LED array |
Wiring the AD7376 potentiometer.
We used the SOIC-16 housing for the AD7376 potentiometer for ease of soldering to wires. The table indicates how each pin was connected. The length of the GND wire from the Arduino board to the shared ground needs to be kept short (2 in. or less) for SPI communication.
| Pin | Connection |
|---|---|
| 1 | +of 10 V power supply |
| 2 | GND (shared GND between that of 10V power supply and Arduino |
| 3 | GND |
| 4 | GND |
| 5 | pin 10 on Arduino (or any other pin designated as a Slave Select, such as 5, 6, or 9 |
| 6 | +5V of Arduino |
| 7 | pin 13 on Arduino (SCLK) |
| 8 | NC (not connected) |
| 9 | NC |
| 10 | NC |
| 11 | pin 11 on Arduino (MOSI) |
| 12 | +5V of Arduino |
| 13 | NC |
| 14 | +of 10V power supply |
| 15 | NC |
| 16 | DIM line of FlexBlock |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32032.043





