TASEP modelling provides a parsimonious explanation for the ability of a single uORF to derepress translation during the integrated stress response
Figures
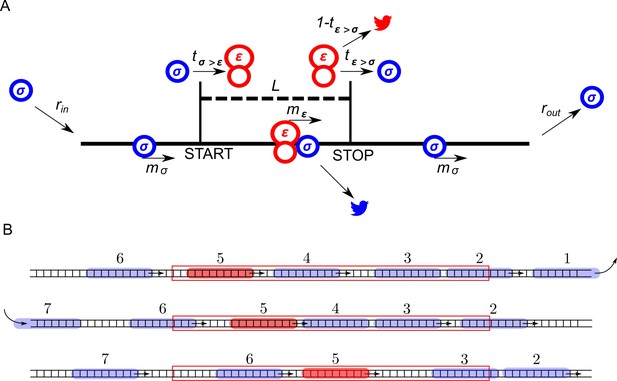
ICIER model.
(A) Parameters of the model. The lattice is shown as a black line with the positions of start and stop codons separated by L sites (codons). Scanning ribosomes are blue and elongating are red in both panels. Ribosome dissociations from mRNAs are indicated with bird symbols. (B) An example of three subsequent states of the lattice with ribosomes shown as semitransparent oval shapes and each ribosome labeled with a unique index. Arrows indicate changes between current and subsequent steps. The red rectangle specifies the location of the uORF.
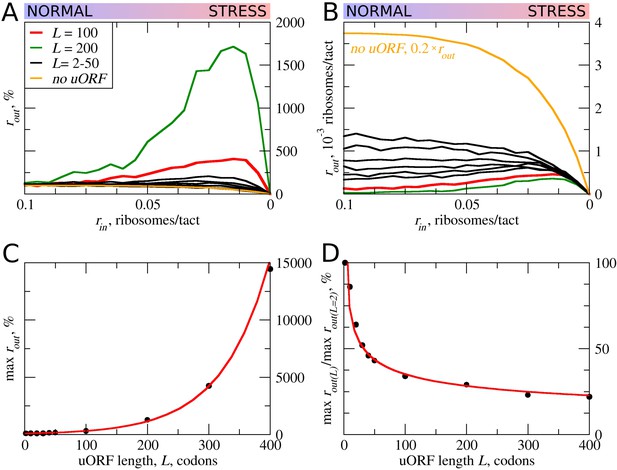
uORF length (L) influences how the flux of scanning ribosomes changes downstream (rout) in response to lowered ribosome load (rin) due to stress.
Other parameters in the simulations: tε>σ=0; tσ>ε=0.8; mσ=mε=0.3. (A, B) Relative (rout[rin]/rout[rin=0.1]) (A) and absolute (B) changes in the density of ribosomes downstream of a uORF for a range of rin from 0.1 to 0 for mRNAs containing uORFs of different lengths (L). (C) Dependence of the relative (rout) maximum on the uORF length. (D) The decrease of maximum rout as a result of increasing uORF length (maximum rout for each L is normalized by rout for L= 2). Source data for panel (A) are provided in Figure 2—source data 1 and for panel (B) in Figure 2—source data 2.
-
Figure 2—source data 1
Source data for Figure 2A.
- https://doi.org/10.7554/eLife.32563.004
-
Figure 2—source data 2
Source data for Figure 2B.
- https://doi.org/10.7554/eLife.32563.005
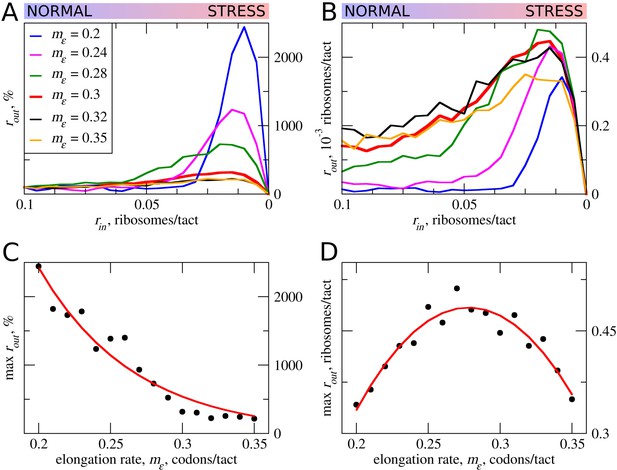
The effect the movement rates (mε) for elongating ribosomes on stress resistance.
Other parameters of the simulations tε>σ=0; tσ>ε=0.8; L=100; mσ=0.3. (A, B) Relative (rout[rin]/rout[rin=0.1]) (A) and absolute (B) changes in the density of ribosomes downstream of the uORF for a range of rin from 0.1 to 0 for different elongating ribosome movement rates (mε)). (C) Dependence of the relative (rout) maximum increase on elongating ribosome movement mε. (D) Dependence of the absolute maximum rout rate on the mε. Source data for panel (A )are provided in Figure 3—source data 1 and for panel (B) in Figure 3—source data 2.
-
Figure 3—source data 1
Source data for Figure 3A.
- https://doi.org/10.7554/eLife.32563.007
-
Figure 3—source data 2
Source data for Figure 3B.
- https://doi.org/10.7554/eLife.32563.008
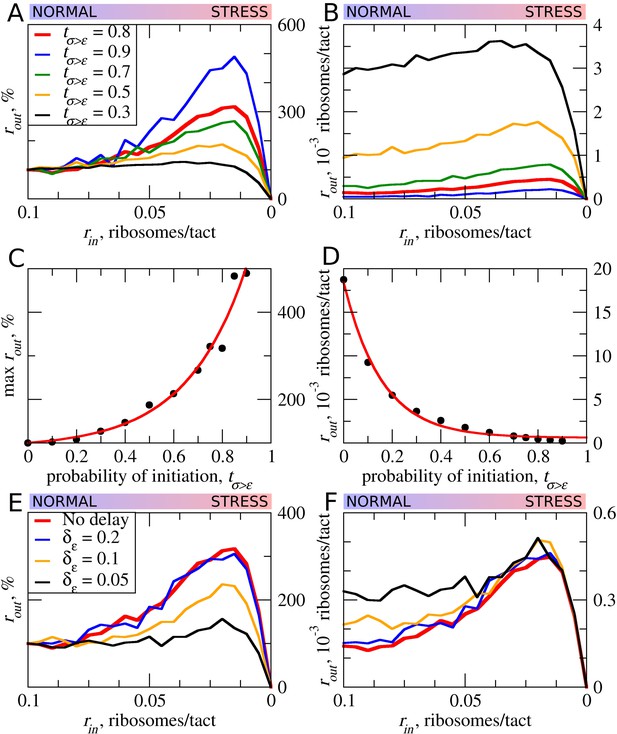
The effect of uORF initiation efficiency (tσ>ε) on stress resistance.
Other parameters of the simulations tε>σ=0; L=100; mσ=0.3; mε=0.3. (A, B) Relative (rout[rin]/rout[rin=0.1]) (A) and absolute (B) changes in the density of ribosomes downstream of a uORF for a range of rin from 0.1 to 0 for different uORF initiation efficiencies (tσ>ε). (C) Dependence of the relative (rout) maximum increase on the uORF initiation efficiency tσ>ε. (D) Dependence of absolute maximum rout rate on tσ>ε. (E, F) Relative (rout[rin]/rout[rin=0.1]) (E) and absolute (F) changes in the density of ribosomes downstream of a uORF depending on the delay δε, which is the probability that newly formed elongating will move. For no delay, δε=1. Source data for panel (A) are provided in Figure 4—source data 1, for panel (B) in Figure 4—source data 2, for panel (E) in Figure 4—source data 3 and for panel (F) in Figure 4—source data 4.
-
Figure 4—source data 1
Source data for Figure 4A.
- https://doi.org/10.7554/eLife.32563.010
-
Figure 4—source data 2
Source data for Figure 4B.
- https://doi.org/10.7554/eLife.32563.011
-
Figure 4—source data 3
Source data for Figure 4E.
- https://doi.org/10.7554/eLife.32563.012
-
Figure 4—source data 4
Source data for Figure 4F.
- https://doi.org/10.7554/eLife.32563.013
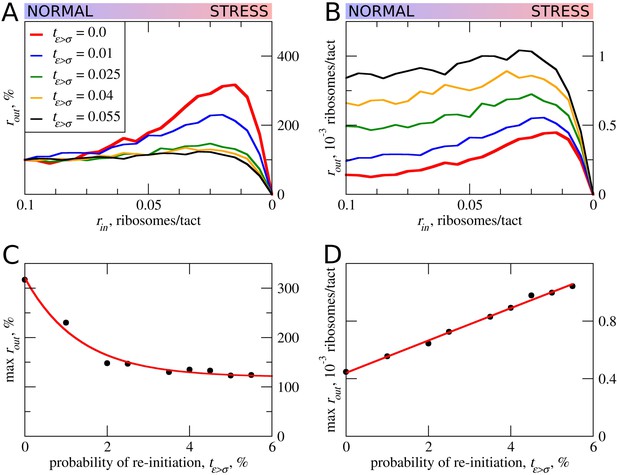
The effect of reinitiation downstream of the uORF on stress resistance.
Other parameters of the simulations tε>σ=0; tσ>ε=0.8; L=100; mσ=0.3; mε=0.3. (A, B) Relative (rout[rin]/rout[rin=0.1]) (A) and absolute (B) changes in the density of ribosomes downstream of a uORF for a range of reinitiation probabilities (tε>σ from 0 to 0.055). (C) Dependence of the relative (rout) maximum increase on the reinitiation probability tε>σ. (D) Dependence of absolute maximum rout rate on the tε>σ. Source data for panel (A) are provided in Figure 5—source data 1 and for panel (B) in Figure 5—source data 2.
-
Figure 5—source data 1
Source data for Figure 5A.
- https://doi.org/10.7554/eLife.32563.015
-
Figure 5—source data 2
Source data for Figure 5B.
- https://doi.org/10.7554/eLife.32563.016
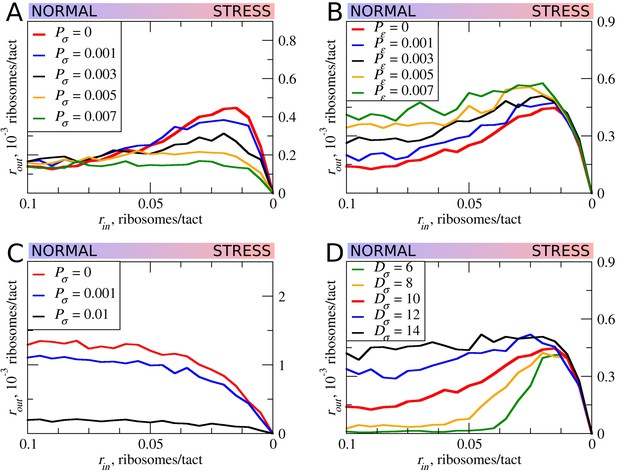
The effect of the size of scanning ribosomes (Dσ) and probability of spontaneous ribosome dissociation (Pσ) on stress resistance.
Other parameters of the simulations; tσ>ε=0.8; L=100; mσ=0.3; mε=0.3. (A, B) Absolute changes in the density of ribosomes downstream of a uORF under a scenario in which scanning (A) or elongating (B) ribosomes could dissociate spontaneously with probabilities Pσ and Pε, respectively. (C) Same as (B) but scanning ribosomes do not dissociate upon collisions with elongating ribosomes. (D) The absolute changes in the densities of ribosomes downstream of a uORF depending on the size of the scanning ribosomes. Source data for panel (A) are provided in Figure 6—source data 1, for panel (B) in Figure 6—source data 2, for panel (C) in Figure 6—source data 3 and for panel (D) in Figure 6—source data 4.
-
Figure 6—source data 1
Source data for Figure 6A.
- https://doi.org/10.7554/eLife.32563.018
-
Figure 6—source data 2
Source data for Figure 6B.
- https://doi.org/10.7554/eLife.32563.019
-
Figure 6—source data 3
Source data for Figure 6C.
- https://doi.org/10.7554/eLife.32563.020
-
Figure 6—source data 4
Source data for Figure 6D.
- https://doi.org/10.7554/eLife.32563.021
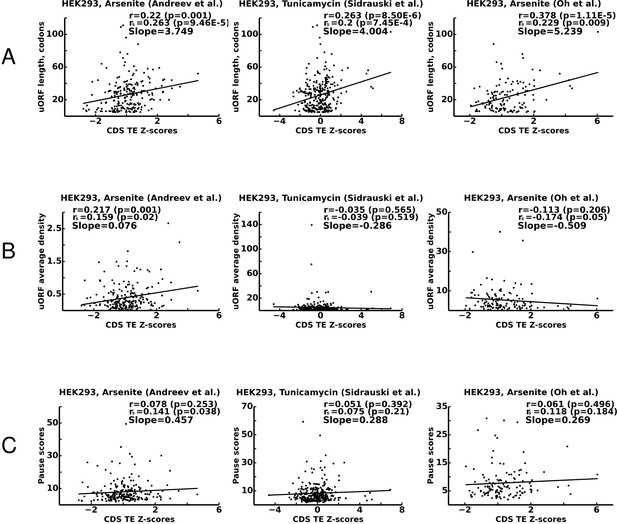
Correlations between translational stress resistance of mRNAs and features in the 5’ leaders of uOFRs for three studies.
(A) uORF length. (B) uORF translation initiation measured as uORF translation efficiency. (C) Ribosome pausing estimated with pause score. Pearson correlation coefficients (r) and Spearman rank correlations (rs) are provided for each plot along with the corresponding p-values. The slope of linear regression is also indicated for each plot. Source data are provided in Figure 7—source data 1.
-
Figure 7—source data 1
- https://doi.org/10.7554/eLife.32563.023
Additional files
-
Supplementary file 1
The parameters of the computer simulations.
- https://doi.org/10.7554/eLife.32563.024
-
Supplementary file 2
The coordinates of the uORFs.
- https://doi.org/10.7554/eLife.32563.025
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32563.026





