COX16 promotes COX2 metallation and assembly during respiratory complex IV biogenesis
Figures
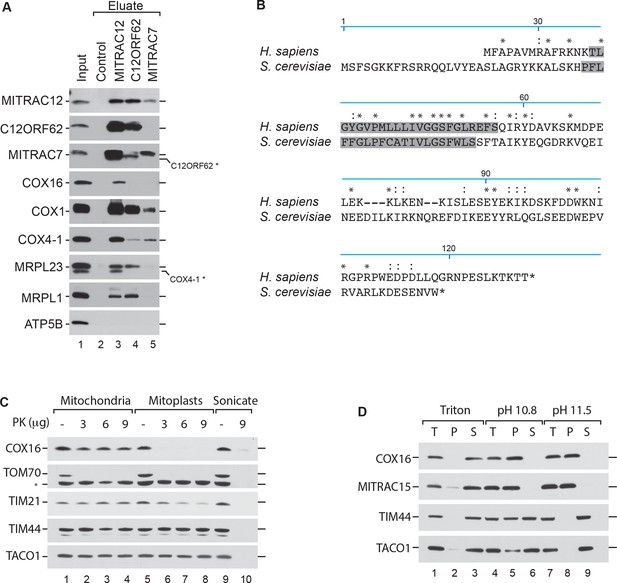
COX16 interacts with the MITRAC complex.
(A) Antibodies against MITRAC12, C12ORF62, MITRAC7, or control antisera were used for immunoisolations of wild type HEK-293T mitochondria. Total, 3%; Eluates, 100%. Residual signals as a result of redeorations are marked in *. (Β) Alignment of the human (H. sapiens) COX16 amino acid sequence to its yeast (S. cerevisiae) homolog using ClustalW. Predicted transmembrane spans are highlighted in gray; asterisk (*) indicates similar residues; colon (:) indicates identical residues. (C) Submitochondrial localization analysis of COX16 using protease protection assays. Wild type and hypotonically swollen mitochondria were treated with proteinase K (PK). Samples were analyzed by SDS-PAGE and western blotting. Asterisk (*), non-specific signal. (D) Membrane association of COX16 was analyzed using mitochondria that were subjected to carbonate extraction or detergent lysis. T, total; S, soluble fraction; P, pellet.
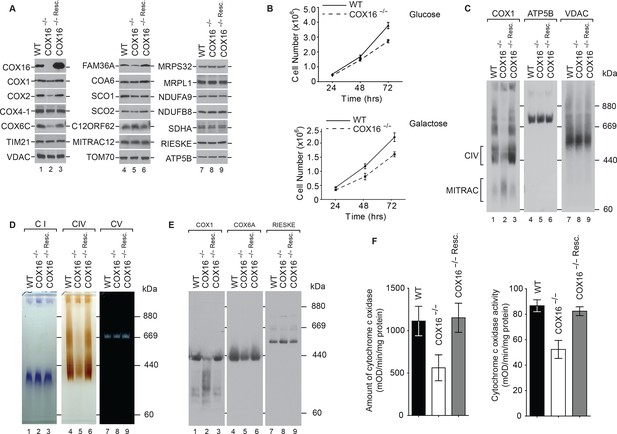
COX16 is required for cytochrome c oxidase biogenesis.
(A) Isolated wild-type (WT), COX16 knockout (COX16–/–) and COX16 knockout expressing WT COX16 from the T-REx locus (COX16–/– Resc.) mitochondria were analyzed by western blotting. (Β) Cell count of wild-type (WT) and COX16 knockout (COX16–/–) grown in medium containing either glucose (left) or galactose (right). (C) Isolated mitochondria from (A), solubilized in 1% Digitonin and analyzed by BN-PAGE and western blotting, subjected with antisera against COX1, ATP5B and VDAC. CIV, complex IV. (D) Isolated mitochondria from (A), solubilized in 1% Digitonin and analyzed by BN-PAGE and in-gel activity assays for complex I (CI), IV (CIV) and V (CV). (E) Isolated mitochondria from (A), were solubilized in 1% N-Dodecyl β-D-maltoside (DDM) and analyzed by BN-PAGE and western blotting with indicated antisera. (F) Measurement of relative amount of cytochrome c oxidase (right) and enzyme activity (left); the mitochondria were isolated as mentioned in (A) (mean ±SEM and n = 3).
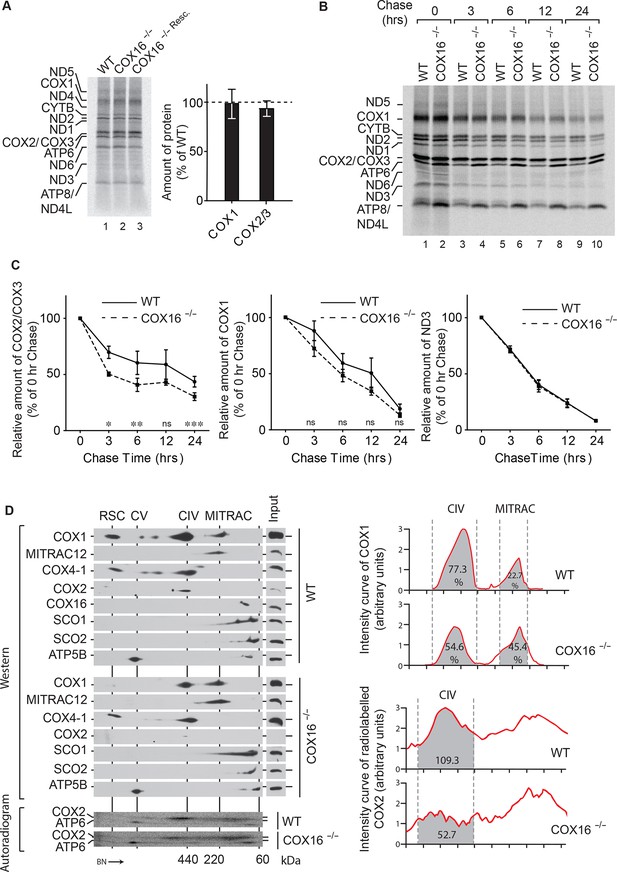
COX16 is required for COX2 assembly.
(A) In vivo labeling of mitochondrial translation products with [35S]methionine in wild-type (WT), COX16 knockout (COX16–/–) and COX16 knockout expressing WT COX16 from the T-REx locus (COX16–/– Resc.). Cells were pulsed for 1 hr and analyzed by SDS-PAGE and digital autoradiography. The values represented are quantifications of the indicated mitochondrial translation products normalized to ND1 (mean ± SEM and n = 3). (Β) Mitochondrial translation products in wild-type (WT) and COX16 knockout (COX16–/–) were labeled with [35S]methionine for 1 hr. Subsequently, the medium was replaced and cells were further cultured in standard medium (chase) for 3, 6, 12 and 24 hr. Cell extracts were analyzed by SDS-PAGE and digital autoradiography. (C) Quantifications using ImageQuant software of the indicated mitochondrial translation products from (Β). The values represented were normalized to ND1 (mean ± SEM and n = 3; *p=0.029, **p=0.042, ***p=0.024, ns = non significant). (D) Protein complexes from wild-type (WT) and COX16 knockout (COX16–/–) mitochondria were extracted under non-denaturing conditions and separated by BN-PAGE, followed by a second dimension SDS-PAGE and western blot analysis (top). Mitochondrial translation products were labeled with [35S]methionine, prior whole cell lysis and complexes separation as described above (bottom). The proteins were detected by using indicated antibodies or by digital autoradiography (COX2, ATP6). Intensity curves (right) for COX1 signals from the western blotting and COX2 from the autoradiogram were calculated using ImageJ. Numbers in the gray regions denote area under intensity curves. For COX1 (top), it is represented as percentage of the total signal in CIV and MITRAC and for COX2 (bottom), as arbitrary units. CIV, Monomeric Complex IV; CV, Complex V; RSC, Respiratory Super-Complexes.
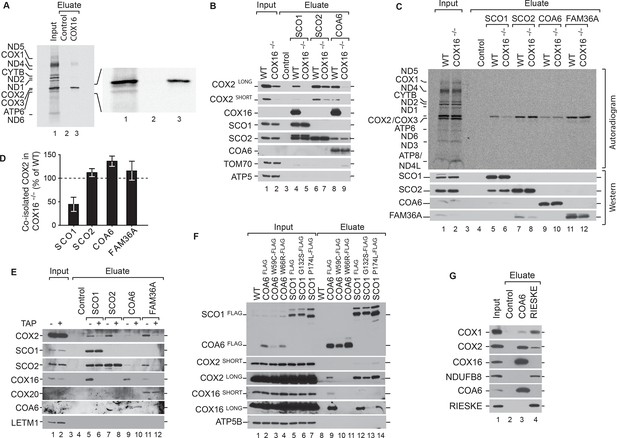
COX16 is required for SCO1 interaction with COX2.
(A) Mitochondrial translation products of wild type cells were labeled with [35S]methionine for 1 hr and whole cell extracts subjected to immunoprecipitation using anti-COX16 or control antisera. The eluates were analyzed by digital autoradiography after SDS-PAGE (Total, 5% and Eluate, 100%). (Β) Immunoprecipitation from wild-type (WT) and COX16 knockout (COX16–/–) mitochondria with anti-SCO1, anti-SCO2, anti-COA6 or control antisera. The eluates were analyzed by western blotting after SDS-PAGE with the indicated antibodies (Total 5% and Eluate, 50%). (C) Mitochondrial translation products in wild-type (WT) and COX16 knockout (COX16–/–) were labeled with [35S]methionine for 1 hr. Whole cell extracts were subjected to immunoprecipitation with anti-SCO1, anti-SCO2, anti-COA6, anti-FAM36A or control antisera. Eluates were separated by SDS-PAGE and Western blotting. Radioactive signals were visualized by digital autoradiography and membranes afterwards decorated with the indicated antibodies. (Total, 5% and Eluate, 100%). (D) Quantification of co-isolated COX2 from immunoprecipitations with the indicated antibodies from (C) (mean ± SEM and n = 3). (E) Mitochondria (without (-) or with thiamphenicol (TAP) (+) treatment) from wild-type (WT) and COX16 knockout cells (COX16–/–) were subjected to immunoprecipitations using antibodies against SCO1, SCO2, COA6 and FAM36A. Samples were subjected to SDS- PAGE and analyzed by western blotting using the indicated antibodies (Total, 5% and Eluate, 100%). (F) Immunoisolations of COA6FLAG or SCO1FLAG along with variants harboring individual pathogenic substitutions (COA6 - W59C and W66R, SCO1 - G132S and P174L). Cells were solubilized and subjected to anti-FLAG immunoprecipitation and eluates analyzed by SDS-PAGE and western blotting using the indicated antibodies. WT, wild type. (Total 5% and Eluate, 50%). (G) Immunoprecipitation from wild-type (WT) mitochondria with anti-COA6, anti-RIESKE or control antisera. The eluates were analyzed by western blotting after SDS-PAGE with the indicated antibodies (total 5% and eluate, 50%).
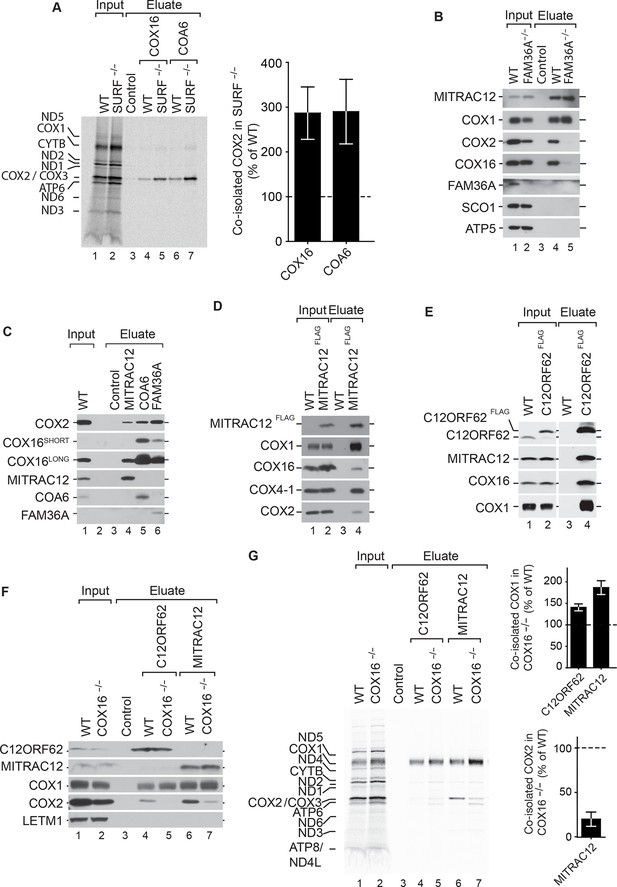
COX16 facilitates integration of COX2 into MITRAC-COX1 modules.
(A) Mitochondrial translation products in wild-type (WT) and SURF knockout (SURF–/–) were labeled with [35S]methionine for 1 hr. Whole cell extracts were subjected to immunoprecipitation using anti-COX16, anti-COA6 or control antisera. Eluates were analyzed by digital autoradiography after SDS-PAGE (Total, 5% and Eluate, 100%). Quantification of co-isolated COX2 amounts with the indicated antibodies were performed using ImageJ (mean ± SEM and n = 3). (Β) Immunoprecipitation from wild-type (WT) and FAM36A knockout (FAM36A–/–) mitochondria with anti-MITRAC12 or control antisera. The eluates were analyzed by western blotting after SDS-PAGE with the indicated antibodies (Total 5% and Eluate, 100%). (C) Immunoprecipitation from wild-type (WT) mitochondria with anti-MITRAC12, anti-COA6, anti-FAM36A or control antisera. The eluates were analyzed by western blotting after SDS-PAGE with the indicated antibodies (Total 5% and Eluate, 50%). (D) Mitochondria isolated from induced MITRAC12FLAG cells were solubilized and subjected to anti-FLAG immunoprecipitation and eluates analyzed by SDS-PAGE and western blotting using the indicated antibodies. WT, wild type. (Total 5% and Eluate, 50%). (E) Mitochondria isolated from induced C12ORF62FLAG cells were solubilized and subjected to anti-FLAG immunoprecipitation and eluates analyzed by SDS-PAGE and western blotting using the indicated antibodies. WT, wild type. (Total 3% and Eluate, 100%). (F) Mitochondria isolated from wild-type (WT) and COX16 knockout (COX16–/–) were used for immunoprecipitation with anti-MITRAC12 or control antisera. The eluates were analyzed by western blotting after SDS-PAGE with the indicated antibodies (Total 5% and Eluate, 50%). (G) Antibodies against C12ORF62, MITRAC12 or control antisera were used for immunoisolation after [35S]methionine labeling of mitochondrial translation products in wild-type (WT) and COX16 knockout (COX16–/–) cells and analyzed by SDS-PAGE and digital autoradiography (Total, 5% and Eluate, 100%). Quantification of co-isolated COX1 or COX2 amounts with the indicated antibodies were performed using ImageJ (mean ± SEM and n = 3).
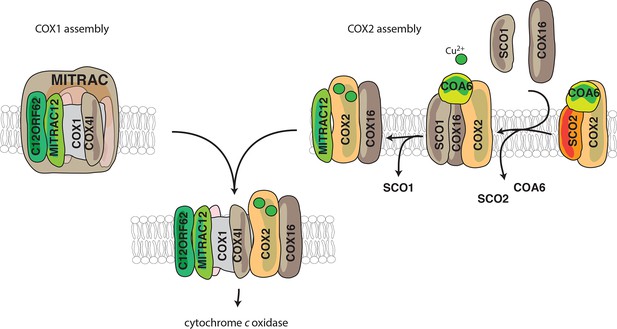
Model for the role of COX16.
COX1 is assembled and guided through the assembly process through its association with MITRAC, where it awaits the association of COX2. COX2 is initially associated with FAM36A and metallochaperones such as SCO2 and COA6 in the early assembly stages. COX16 acts at the later stages of COX2 assembly. The early assembly factors are apparently no longer associated with the complex at this stage. COX16 facilitates the association of SCO1 and thus probably leads to proper COX2 maturation. It then facilitates the merger of COX1 and COX2 assembly lines after the exit of SCO1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | COX16 | NCBI | NCBI Gene ID: 51241 | First study to adress Human COX16 |
| Cell line (Homo sapiens) | HEK293-Flp-InTM T-RexTM (HEK293T) Cell Line | ThermoFisher Scientific | RRID:CVCL_U421 | |
| Cell line (Homo sapiens) | HEK293-Flp-InTM T-RexTM (HEK293T)-COX16-/- | This paper | N/A | Cell line generated as described in Materials and methods |
| Cell line (Homo sapiens) | HEK293-Flp-InTM T-RexTM (HEK293T)-COX16-/- COX16 OE | This paper | N/A | Cell line generated as described in Materials and methods |
| Transfected construct (Homo sapiens) | pX330-COX16 gRNA | This paper | N/A | cloning described in Materials and methods |
| Transfected construct (Homo sapiens) | pEGFPN1 | Clonetech | N/A | |
| Transfected construct (Homo sapiens) | pCDNA5-COX16 | This paper | N/A | Construct generated from amplifying COX16 from WT HEK cDNA |
| Transfected construct (Homo sapiens) | pCDNA5-COA6-FLAG | This paper | N/A | Construct generated from amplifying COA6 from WT HEK cDNA |
| Transfected construct (Homo sapiens) | pCDNA5-COA6-W59C-FLAG | This paper | N/A | Construct generated by mutagenesis of pCDNA5-COA6-FLAG |
| Transfected construct (Homo sapiens) | pCDNA5-COA6-W66R-FLAG | This paper | N/A | Construct generated by mutagenesis of pCDNA5-COA6-FLAG |
| Transfected construct (Homo sapiens) | pCDNA5-SCO1 | This paper | N/A | Construct generated from amplifying SCO1 from WT HEK cDNA |
| Transfected construct (Homo sapiens) | pCDNA5-SCO1-G132S-FLAG | This paper | N/A | Construct generated by mutagenesis of pCDNA5-SCO1-FLAG |
| Transfected construct (Homo sapiens) | pCDNA5-SCO1-P174L-FLAG | This paper | N/A | Construct generated by mutagenesis of pCDNA5-SCO1-FLAG |
| Biological sample () | N/A | |||
| Antibody | MITRAC12 | self made | PRAB3761 | (1:1000) |
| Antibody | C12ORF62 | self made | PRAB4844 | (1:500) |
| Antibody | MITRAC7 | self made | PRAB4843 | (1:500) |
| Antibody | COX16 | Proteintech | RRID:AB_10666854 | (1:1000) |
| Antibody | COX1 | self made | PRAB2035 | (1:2000) |
| Antibody | COX4-1 | self made | PRAB1522 | (1:2000) |
| Antibody | MRPL23 | self made | PRAB1716 | (1:500) |
| Antibody | MRPL1 | self made | PRAB4969 | (1:500) |
| Antibody | TOM70 | self made | PRAB3280 | (1:1000) |
| Antibody | TACO1 | self made | PRAB3627 | (1:500) |
| Antibody | MITRAC15 | self made | PRAB4814 | (1:500) |
| Antibody | FLAG | Sigma Aldrich | RRID:AB_259529 | (1:2000) |
| Antibody | COX2 | Abcam | ab110258 | (1:2000) |
| Antibody | TIM21 | self made | PRAB3674 | (1:2000) |
| Antibody | VDAC | self made | PRAB1515 | (1:1500) |
| Antibody | SDHA | self made | PRAB4978 | (1:2000) |
| Antibody | Rieske | self made | PRAB1512 | (1:2000) |
| Antibody | ATP5B | self made | PRAB4826 | (1:10000) |
| Antibody | TIM44 | Proteintech | RRID:AB_2204679 | (1:2500) |
| Antibody | NDUFB8 | self made | PRAB3764 | (1:500) |
| Antibody | NDUFA9 | self made | PRAB1524 | (1:500) |
| Antibody | LETM1 | self made | PRAB538 | (1:5000) |
| Antibody | TIM23 | self made | PRAB1527 | (1:2000) |
| Antibody | SCO1 | self made | PRAB4980 | (1:500) |
| Antibody | COA6 | self made | PRAB5007 | (1:500) |
| Antibody | COX6C | self made | PRAB4950 | (1:2000) |
| Antibody | SCO2 | self made | PRAB4982 | (1:500) |
| Antibody | FAM36A | self made | PRAB4490 | (1:500) |
| Antibody | SURF1 | self made | PRAB1528 | (1:1000) |
| Recombinant DNA reagent | QuikChange Site-Directed Mutagenesis Kit | Agilent | 210515 | |
| Recombinant DNA reagent | KOD Hot Start DNA Polymerase | Merck | 71086–3 | |
| Recombinant DNA reagent | First Strand cDNA Synthesis kit | ThermoFisher Scientific | K1612 | |
| Commercial assay or kit | Human Complex IV activity kit | Abcam | ab109910 | |
| Chemical compound, drug | GeneJuice | Merck | 70967–3 | |
| Chemical compound, drug | Anti-FLAG M2 Affinity Gel | Sigma-Aldrich | A2220 | |
| Chemical compound, drug | Trizol | ThermoFisher Scientific | 15596026 | |
| Chemical compound, drug | Protein-A SepharoseTM CL-4B | GE Healthcare | 17-0963-03 | |
| Chemical compound, drug | [35S]methionine | Hartmann Analytic | SCM-01 | |
| Chemical compound, drug | Emetine dihydrochloride hydrate | Sigma-Aldrich | 219282 | |
| Chemical compound, drug | Anisomycin | AppliChem | A7650,0025 | |
| Software, algorithm | ImageQuantTL 7.0 software | GE Healthcare | RRID:SCR_014246 | |
| Software, algorithm | ImageJ 1.47 v | NIH | RRID:SCR_003070 | |
| Software, algorithm | Geneious | Biomatters Ltd | RRID:SCR_010519 | |
| Software, algorithm | Prism5 | GraphPad Software | RRID:SCR_015807 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32572.008





