Ecological multiplex interactions determine the role of species for parasite spread amplification
Figures
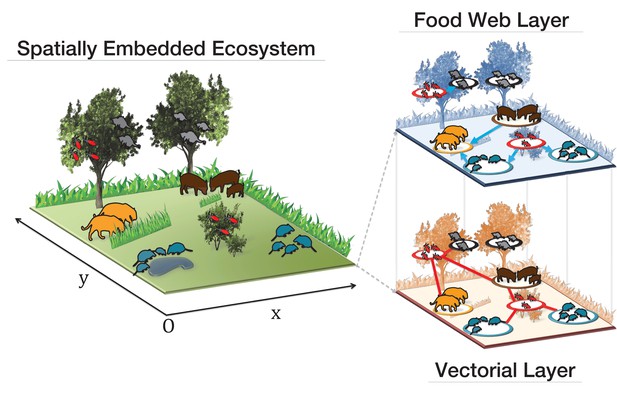
Visual representation of our ecological multiplex network model.
The background colouring on the right panels indicates that elements such as bushes, trees, grass and water areas are not present in the spatial embedding of the ecomultiplex model, where only the spatial network structure among close animal groups is considered.
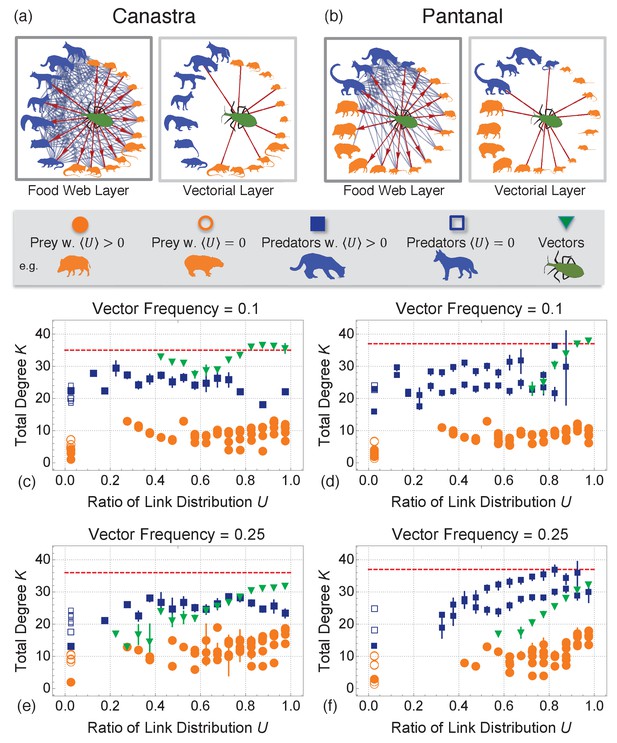
Single layer interactions and multiplex cartographies of Canastra and Pantanal biomes.
(a) and (b) Food web layer and vectorial layer in Canastra (left) and Pantanal (right) biomes. Predators are highlighted in blue, preys in orange and the vectors in green. Interactions involving the insect are highlighted in red. Interactions involving other species are reported for completeness in blue. (c–f): Multiplex cartography of the Canastra ecomultiplex network with 10% (c) and 25% (e) of total groups as vectors. Multiplex cartography of the Pantanal ecomultiplex network with 10% (d) and 25% (f) of total groups as vectors. The red line separates hub nodes, i.e. the most connected nodes within the 95th percentile of the total degree distribution. The cartography highlights the average trends of species: blue for predators, orange for preys, and green for vectors. As evident from (a–e), vectors have higher total degree in the ecosystem and tend to distribute more equally their links across both the multiplex layers than all other species. Vectors are therefore pivotal in the ecosystem.
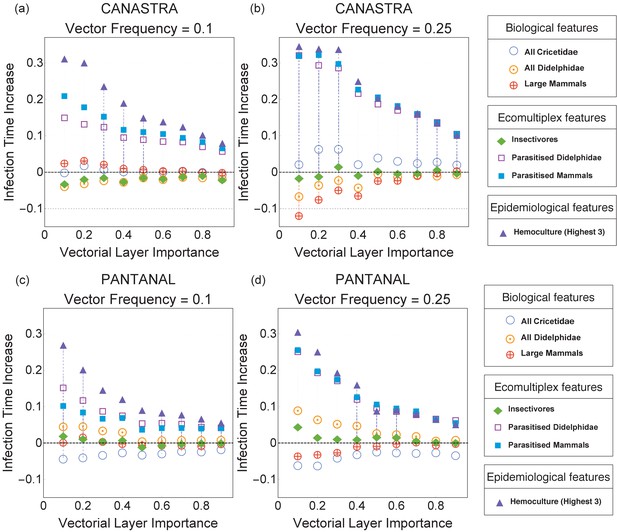
Immunisation strategies for the Canastra (top) and Pantanal (bottom) ecosystems when the vector frequency is 0.1 (left) and 0.25 (right).
The infection time increase plotted on the y-axis is defined in Immunisation Strategies (Methods and Materials). An increase of +0.3 indicates that the infection time in a given immunisation scenario was 30% higher than in the reference case of random immunisation.
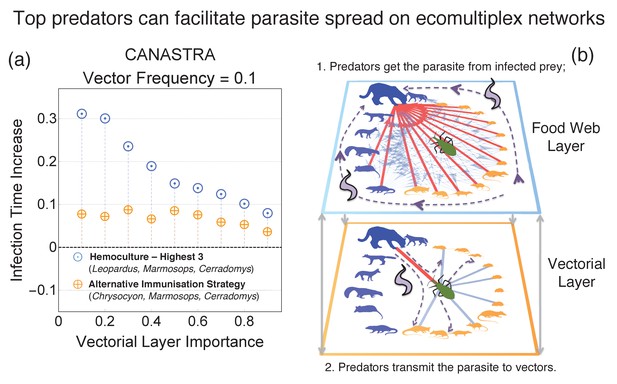
Difference in performances of the best immunisation strategy (hemoculture - Highest 3) when another top predator in the ecosystem (not interacting with the vector in the vectorial layer) is immunised instead of Leopardus pardalis (hemoculture - H 3 No Leopardus).
The other top predator is the maned wolf (Chrysocyon brachiurus).
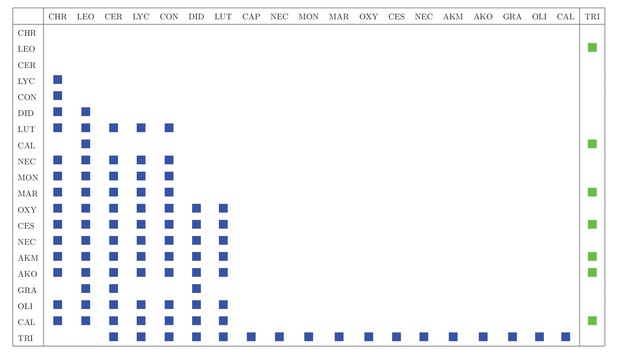
Interaction matrix includes trophic (predator-prey, blue squares) and vectorial (vector-host, green squares) interactions in Canastra area.
https://doi.org/10.7554/eLife.32814.010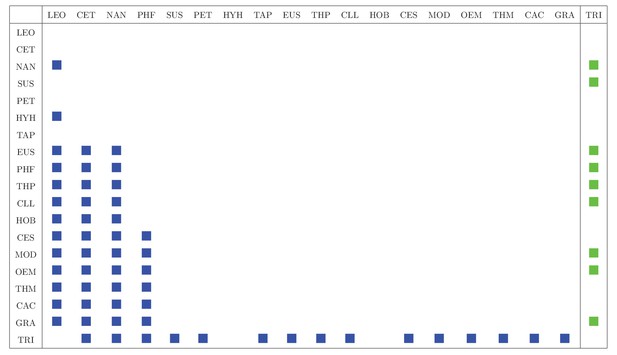
Interaction matrix includes trophic (predator-prey, blue squares) and vectorial (vector-host, green squares) interactions in Pantanal area.
https://doi.org/10.7554/eLife.32814.011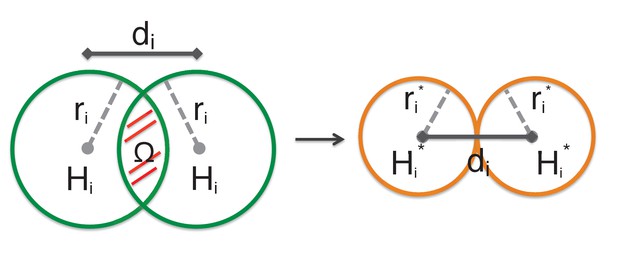
In general home ranges for different animal groups of the same species can overlap (e.g.
see the red overlapping area on the left). Hence, not all the energy available in the home range can sustain the biomass of the animal groups as some resources must be shared and competition can occur. We therefore approximate the total energy effectively available from the overlapping home ranges with the energy coming from non-overlapping effective home ranges , which allow to approximate the total biomass for species . Effective home ranges have radius equal to half the distance between animal groups. .
Scheme on which species are immunised as animal groups in the ecomultiplex network in the different immunisation strategies presented in the main text.
The average frequency, serology and hematology of the animal groups immunised in each strategy are presented as well. Error margins indicate standard deviations.
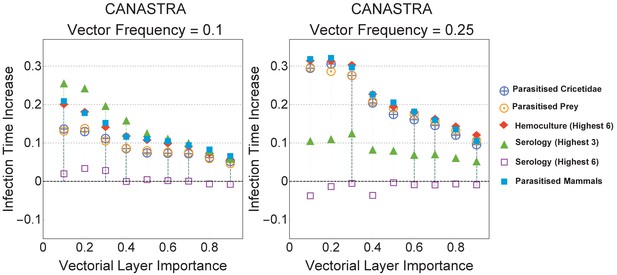
Infection time increases in Canastra for immunisation strategies that are not reported in the main text.
Visual comparisons are made against the strategy Parasitised Mammals from the main text. For low vector frequency () all the reported strategies behave worse than Hemoculture (Highest 3) and were therefore not discussed in the main text.
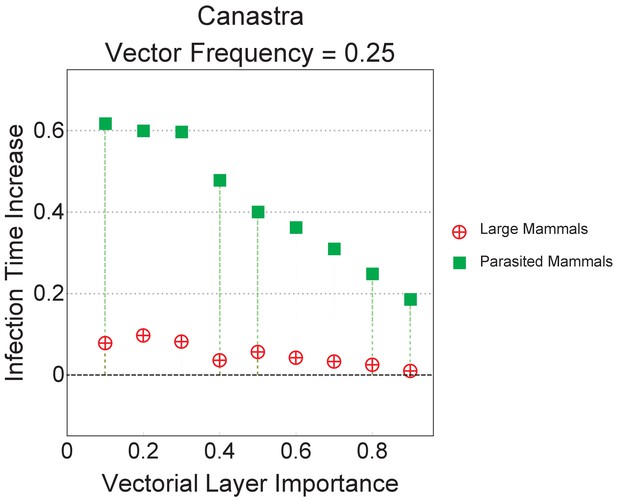
The best and the worst performing strategies presented in the main text for the Canastra ecosystem are here presented relatively to the null model with equal abundances.
In this null model we consider an ecomultiplex network where all animal groups have equal abundance, that is, occur with equal frequency. The error margins in the plot are the same size of the dots and are based on 500 iterations. Even providing equal abundances to different species does not remove the gap in global infection time that was observed in the main text.
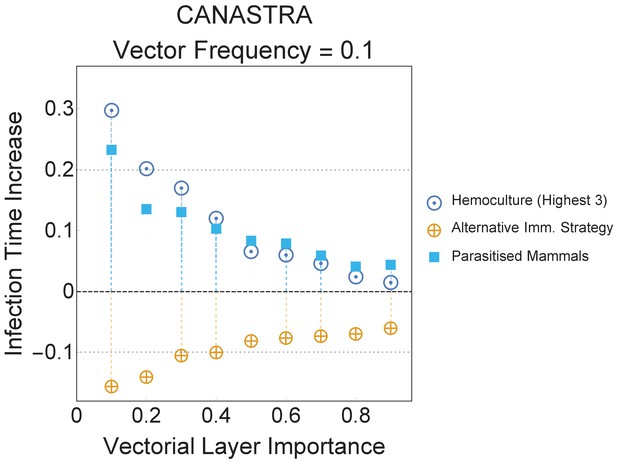
The best network-based and the worst performing strategies presented in the main text for the Canastra ecosystem are here presented relatively to the null model with equal abundances.
In this null model we consider an ecomultiplex network where all animal groups have equal abundance, that is, occur with equal frequency. The error margins in the plot are the same size of the dots and are based on 500 iterations. Even providing equal abundances to different species does not remove the gap in global infection time that was observed in the main text.
Tables
Immunisation types, names and targets of the strategies we tested (Appendix 1).
https://doi.org/10.7554/eLife.32814.007| Immunisation type | Strategy name | Strategy targets |
|---|---|---|
| Ecomultiplex Topological Features | Insectivores | Species feeding on the vector in a food-web |
| Parasitised Didelphidae | Didelphidae contaminated by the vector on a vectorial layer | |
| Parasitised Mammals | All species contaminated by the vector on a vectorial layer | |
| Taxonomic/morphological features | All Cricetidae | All Cricetidae |
| All Didelphidae | All Didelphidae | |
| Large Mammals | All species with a body mass > 1 kg | |
| Epidemiological Features | Hemoculture N | The species with the highest likelihood of being found infected with the parasite in field work (see Appendix 1). |
| Serology N | The species with the highest likelihood of having been infected with the parasite during their life time (see Appendix 1). |
Taxonomic and ecological data of different animal species in Canastra area (1- (Reis et al., 2006), 2- (Myers et al., 2008), 3- (Herrera et al., 2011), 4- (Bonvicino et al., 2008), 5- (Schofield, 1994)).
(Rocha et al., 2013) report prevalence measurements for some genera, indicating the species that were collected during the study. Because those species are ecologically similar we calculated the average body size of the genus considering species that were collected. M. americana, M. domestica and M. sorex. A. sp., A. lindberghi, A. cursor. O. sp, O. nigripes, O. rupestris. C. sp, C. tener.
| ID | Species | Common name | Family | Diet type | Biomass [gr] | References |
|---|---|---|---|---|---|---|
| CHR | Chrysocyon brachyurus | Maned wolf | Canidae | omnivorous | 25000 | [1, 2] |
| LEO | Leopardus pardalis | Ocelot | Felidae | carnivorous | 9740 | [1, 2, 3] |
| CER | Cerdocyon thous | Crab-eating fox | Canidae | omnivorous | 6600 | [1, 2, 3] |
| LYC | Lycalopex vetulus | Hoary fox | Canidae | omnivorous | 3350 | [1, 2] |
| CON | Conepatus semistriatus | Striped˙hog-nosed skunk | Mustelidae | omnivorous | 2567 | [1, 2] |
| DID | Didelphis albiventris | White-eared opossum | Didelphidae | omnivorous | 1625 | [1, 2] |
| LUT | Lutreolina crassicaudata | Thick-tailed opossum | Didelphidae | omnivorous | 600 | [1, 2] |
| CAP | Caluromys philander | Bare-tailed˙woolly opossum | Didelphidae | omnivorous | 255 | [1, 2] |
| NEC | Nectomys squamipes | South American water rat | Cricetidae | omnivorous | 270 | [1, 2] |
| MON | Monodelphis spp a | Short-tailed opossum | Didelphidae | omnivorous | 72.8 | [1, 2, 3] |
| MAR | Marmosops incanus | Gray slander opossum | Didelphidae | omnivorous | 80 | [1, 2] |
| OXY | Oxymycterus delator | Spy hocicudo | Cricetidae | omnivorous | 78.3 | [4] |
| CES | Cerradomys subflavus | Rice rat | Cricetidae | omnivorous | 73 | [4] |
| NEC | Necromys lasiurus | Hairy-tailed bolo mouse | Cricetidae | omnivorous | 52 | [1, 2, 4] |
| AKM | Akodon montensis | Montane grass mouse | Cricetidae | omnivorous | 40.5 | [1, 2, 4] |
| AKO | Akodon spp b | Grass mouse | Cricetidae | omnivorous | 33.25 | [1, 4] |
| GRA | Gracilinanus agilis | Agile gracile opossum | Didelphidae | omnivorous | 27.25 | [1, 2, 3] |
| OLI | Oligoryzomys spp c | Pygmy rice rats | Cricetidae | omnivorous | 21.23 | [1, 4] |
| CAL | Calomys spp d | Vesper mouse | Cricetidae | herbivorous | 18.65 | [1, 4] |
| TRI | Triatominae | Kissing bug | Reduviidae | blood | 0.2 | [5] |
Taxonomic and ecological data of different animal species in Pantanal area (1- (Reis et al., 2006), 2- (Myers et al., 2008), 3- (Herrera et al., 2011), 4- (Bonvicino et al., 2008), 5- (Schofield, 1994)).
https://doi.org/10.7554/eLife.32814.013| ID | Species | Common name | Family | Diet type | Biomass [gr] | References |
|---|---|---|---|---|---|---|
| LEO | Leopardus pardalis | Ocelot | Felidae | carnivorous | 9740 | [1, 2, 3] |
| CET | Cerdocyon thous | Crab-eating fox | Canidae | omnivorous | 6600 | [1, 2, 3] |
| NAN | Nasua nasua | South American coati | Procyonidae | omnivorous | 5140 | [1, 2, 3] |
| SUS | Sus scrofa | Wild boar | Suidae | omnivorous | 105750 | [1, 2, 3] |
| PET | Pecari tajacu | Collared peccary | Tayassuidae | omnivorous | 24000 | [1, 2, 3] |
| HYH | Hydrochaeris hydrochaeris | Capybara | Caviidae | herbivorous | 46200 | [1, 2, 3] |
| TAP | Tayassu pecari | White-lipped peccary | Tayassuidae | omnivorous | 30200 | [1, 2, 3] |
| EUS | Euphractus sexcinctus | Six-banded armadillo | Chlamyphoridae | omnivorous | 4450 | [1, 2, 3] |
| PHF | Philander frenatus | Southeastern four-eyed opossum | Didelphidae | omnivorous | 306.2 | [1, 2, 3] |
| THP | Thrichomys pachyurus | Paraguayan punar | Echimyidae | herbivorous | 291.25 | [1, 3, 4] |
| CLL | Clyomys laticeps | Broad-headed spiny rat | Cricetidae | herbivorous | 187.33 | [1, 3, 4] |
| HOB | Holochilus brasiliensis | Web-footed marsh rat | Cricetidae | herbivorous | 196 | [1, 3, 4] |
| CES | Cerradomys scotti | Lindbergh’s rice rat | Cricetidae | omnivorous | 92.34 | [3, 4] |
| MOD | Monodelphis domestica | Gray short-tailed opossum | Didelphidae | omnivorous | 111.2 | [1, 2, 3] |
| OEM | Oecomys mamorae | Mamore arboreal rice rat | Cricetidae | herbivorous | 83.75 | [1, 3, 4] |
| THM | Thylamys macrurus | Long-tailed fat-tailed opossum | Didelphidae | omnivorous | 46.6 | [1, 2, 3] |
| CAC | Calomys callosus | Large vesper mouse | Cricetidae | herbivorous | 37.45 | [1, 3] |
| GRA | Gracilinanus agilis | Agile gracile opposum | Didelphidae | omnivorous | 27.25 | [1, 2, 3] |
| TRI | Triatominae | Kissing bug | Reduviidae | blood | 0.2 | [5] |
Scheme on which species are immunised as animal groups in the ecomultiplex network in the different immunisation strategies presented in the main text.
The average frequency, serology and hematology of the animal groups immunised in each strategy are presented as well. Error margins indicate standard deviations.
| CANASTRA | Immune and Susceptible Species in the Immunisation Strategies ↓ | ||||||
|---|---|---|---|---|---|---|---|
| Species ↓ | All Cricetidae | All Didelphidae | Large Mammals | Parasitised Mammals | Parasitised Didelphidae | Insectivores | Hemoculture 3 |
| Chrysocyon brachyurus | |||||||
| Leopardus pardalis | |||||||
| Cerdocyon thous | |||||||
| Lycalopex vetulus | |||||||
| Conepatus semistriatus | |||||||
| Didelphis albiventris | |||||||
| Lutreolina crassicaudata | |||||||
| Caluromys philander | |||||||
| Nectomys squamipes | |||||||
| Monodelphis spp | |||||||
| Marmosops incanus | |||||||
| Oxymycterus delator | |||||||
| Cerradomys subflavus | |||||||
| Necromys lasiurus | |||||||
| Akodon montensis | |||||||
| Akodon spp | |||||||
| Gracilinanus agilis | |||||||
| Oligoryzomys spp | |||||||
| Calomysspp | |||||||
| Mean Frequency (fv = 0.1) | 640 ± 50 | 480 ± 70 | 200 ± 20 | 560 ± 80 | 520 ± 50 | 510 ± 50 | 400 ± 100 |
| Mean Frequency (fv = 0.25) | 540 ± 40 | 400 ± 60 | 170 ± 20 | 470 ± 70 | 430 ± 40 | 430 ± 40 | 400 ± 100 |
| Mean Serology | 0.02 ± 0.02 | 0.2 ± 0.1 | 0.3 ± 0.2 | 0.3 ± 0.2 | 0.3 ± 0.3 | 0.12 ± 0.07 | 0.7 ± 0.3 |
| Mean Hemoculture | 0.07 ± 0.04 | 0.1 ± 0.1 | 0.2 ± 0.2 | 0.3 ± 0.1 | 0.3 ± 0.2 | 0.07 ± 0.04 | 0.6 ± 0.2 |
| PANTANAL | Immune and Susceptible Species in the Immunisation Strategies ↓ | ||||||
|---|---|---|---|---|---|---|---|
| Species ↓ | All Cricetidae | All Didelphidae | Large Mammals | Parasitised Mammals | Parasitised Didelphidae | Insectivores | Hemoculture 3 |
| Leopardus pardalis | |||||||
| Cerdocyon thous | |||||||
| Nasua nasua | |||||||
| Sus scrofa | |||||||
| Pecari tajacu | |||||||
| Hydrochaeris hydrochaeris | |||||||
| Tayas supecari | |||||||
| Euphractus sexcinctus | |||||||
| Philander frenatus | |||||||
| Thrichomys pachyurus | |||||||
| Clyomys laticeps | |||||||
| Holochilus brasiliensis | |||||||
| Cerradomys scotti | |||||||
| Monodelphis domestica | |||||||
| Oecomys mamorae | |||||||
| Thylamys macrurus | |||||||
| Calomys callosus | |||||||
| Gracilinanus agilis | |||||||
| Mean Frequency (fv = 0.1) | 730 ± 60 | 800 ± 100 | 210 ± 20 | 540 ± 90 | 700 ± 100 | 530 ± 80 | 600 ± 200 |
| Mean Frequency (fv = 0.1) | 610 ± 50 | 650 ± 80 | 170 ± 20 | 450 ± 80 | 630 ± 100 | 440 ± 60 | 500 ± 200 |
| Mean Serology | 0.24 ± 0.02 | 0.5 ± 0.1 | 0.3 ± 0.1 | 0.37 ± 0.09 | 0.5 ± 0.1 | 0.35 ± 0.06 | 0.50 ± 0.09 |
| Mean Hemoculture | 0.02 ± 0.01 | 0.18 ± 0.06 | 0.04 ± 0.03 | 0.12 ± 0.3 | 0.23 ± 0.03 | 0.07 ± 0.02 | 0.25 ± 0.02 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.32814.008





