Presenilin mutations deregulate mitochondrial Ca2+ homeostasis and metabolic activity causing neurodegeneration in Caenorhabditis elegans
Figures
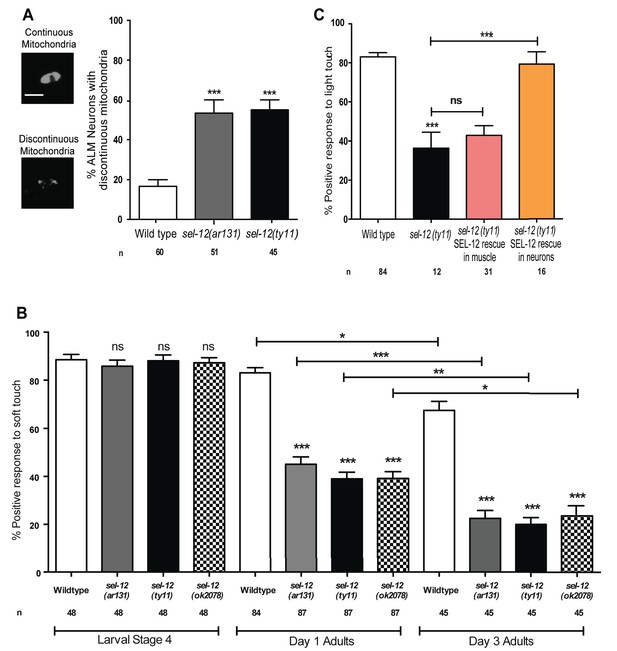
SEL-12 is required for mitochondrial structural maintenance and mechanosensation.
(A) Representative images and quantification of the incidence of discontinuous ALM neuronal mitochondria. Analysis was done using transgenic animals expressing mCherry fused with the outer mitochondrial membrane protein, TOMM-20 in mechanosensory neurons (twnEx8). Scale bar represents 10 μm. (B) Response of wild type and sel-12 mutants to anterior and posterior light touch at larval stage 4, day 1 and 3 of adulthood. (C) Response of day one sel-12(ty11) animals with tissue specific SEL-12 expression to light touch. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, *p<0.05, **p<0.001, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 2—source data 1
Raw data for Figure 2.
- https://doi.org/10.7554/eLife.33052.004
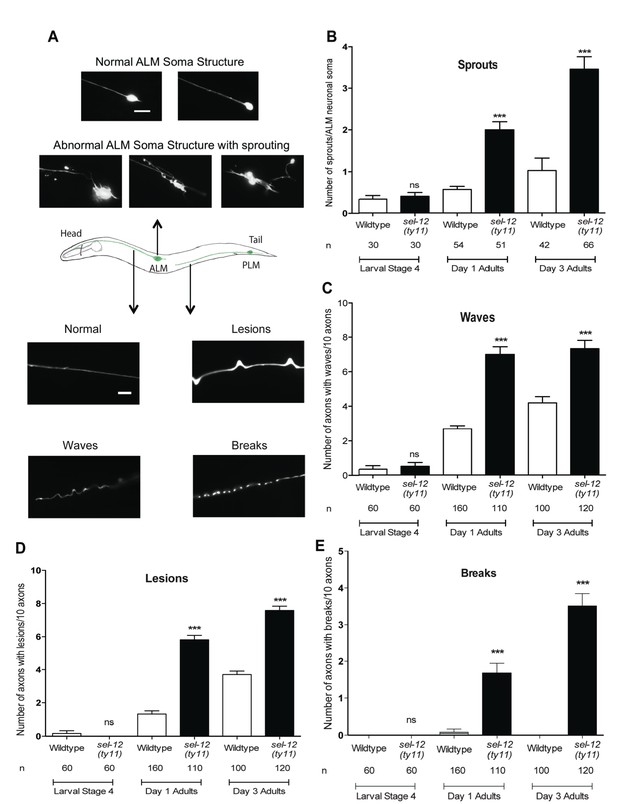
Loss of SEL-12 function results in neurodegeneration.
(A) Representative images of normal ALM neuronal soma and abnormal soma with ectopic sprouting (above), cartoon depicting location of ALM and PLM neuron in C. elegans (middle) and representative images of wave-like processes, lesions and breaks observed in the ALM and PLM neuronal processes (below). Scale bar represents 10 μm. (B) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma at larval stage 4, day 1 and 3 of adulthood. Quantification of the frequency of wave-like processes (C), lesions (D) and breaks (E) in ALM and PLM neuronal processes in wild type and sel-12(ty11) animals at larval stage 4, day 1 and 3 of adulthood. Neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 3—source data 1
Raw data for Figure 3.
- https://doi.org/10.7554/eLife.33052.007
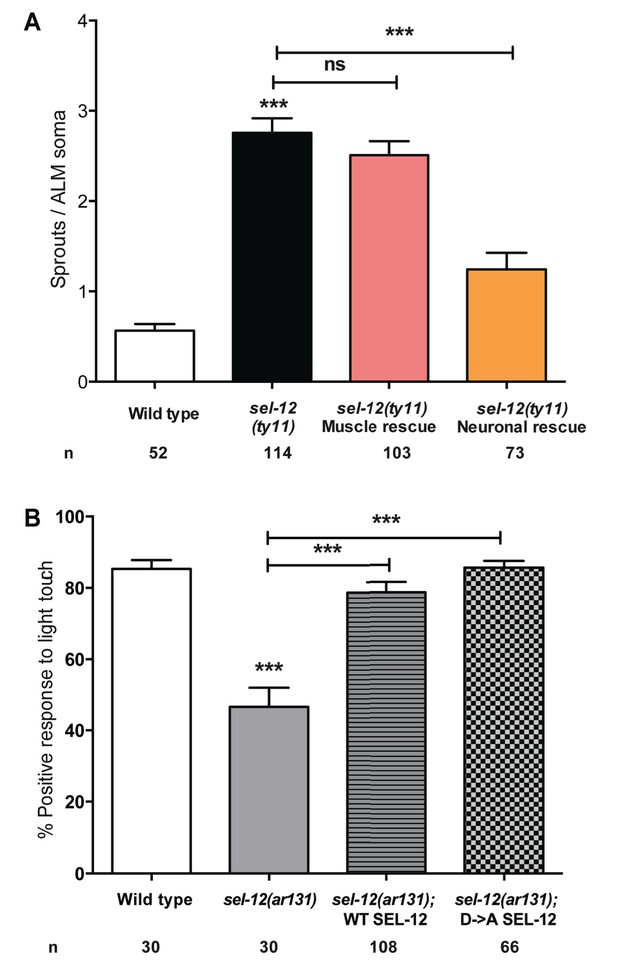
SEL-12 has a tissue-specific but protease independent role in neuronal health maintenance.
(A) Quantification of aberrant structures (sprouts/branches) in ALM neurons of day one sel-12(ty11) animals with tissue specific SEL-12 expression. ALM neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). (B) Quantification of the response of wild type, sel-12(ar131) and sel-12(ar131) animals expressing wild type or a protease dead SEL-12 to anterior and posterior light touches. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 3—figure supplement 1—source data 1
Raw data for Figure 3—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.008
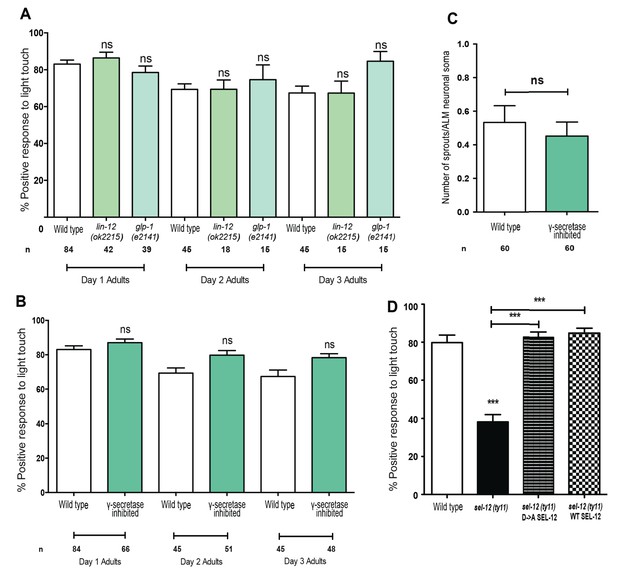
Notch signaling and gamma-secretase activity is not required for mechanosensation or mechanosensory neuron morphology.
(A) Quantification of the response of Notch mutants (glp-1 and lin-12 mutants) to light anterior and posterior touch at day 1, 2 and 3 of adulthood. (B) Quantification of response to light touch in animals treated with gamma-secretase inhibitor (Compound E). (C) Quantification of aberrant neuronal structures (sprouts/branches) in ALM neurons in day one adult animals treated with gamma-secretase inhibitor (Compound E). (D) Quantification of the response of wild type, sel-12(ty11) and sel-12(ty11) animals expressing wild type or a protease dead SEL-12 to anterior and posterior light touch. ALM neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). n = number of animals examined per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey test for A,B, and D, two-tailed T-test for C).
-
Figure 4—source data 1
Raw data for Figure 4.
- https://doi.org/10.7554/eLife.33052.011
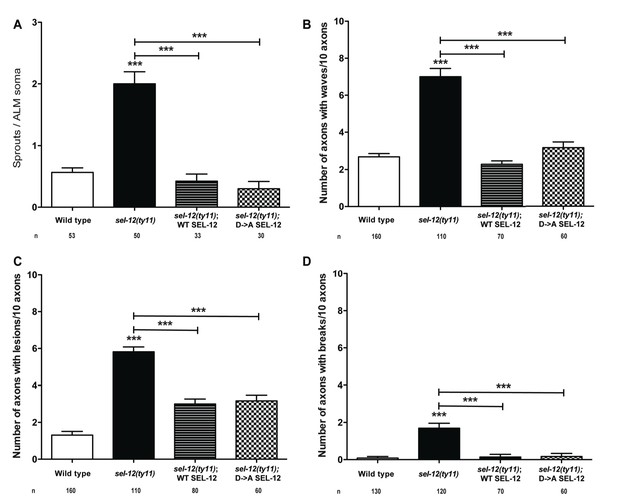
Loss of gamma-secretase function does not result in neurodegenerative morphologies.
(A) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma of wild type, sel-12(ty11) and sel-12(ty11) animals expressing wild type or a protease dead SEL-12 at day 1 of adulthood. Quantification of wave-like processes (B), lesions (C) and breaks (D) in ALM and PLM neuronal processes of wild type, sel-12(ty11) and sel-12(ty11) animals expressing wild type or a protease dead SEL-12 at day 1 of adulthood. Neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 4—figure supplement 1—source data 1
Raw data for Figure 4—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.012
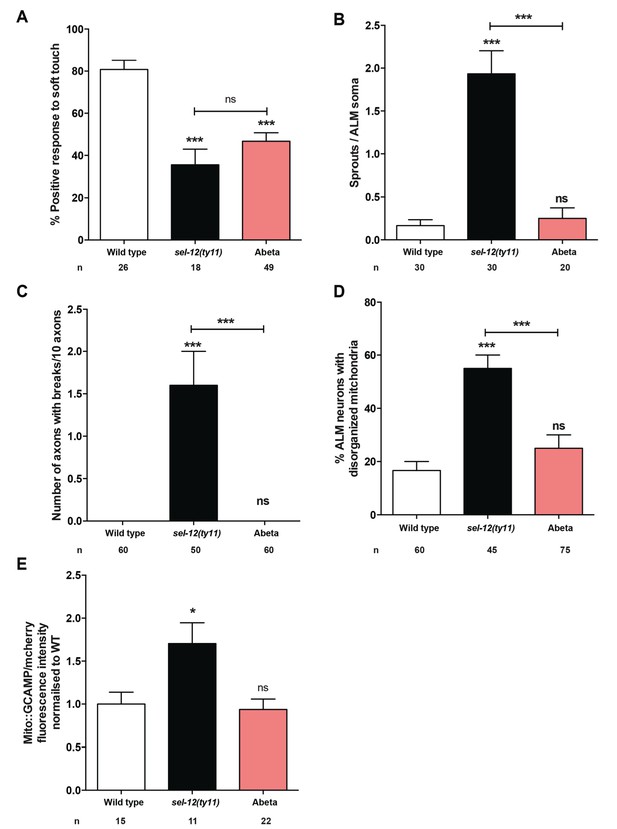
Expression of human Abeta1-42 peptide in the the nervous system results in functional loss dissimilar from sel-12 mutants.
(A) Quantification of the response of Abeta peptide expressing animals to light anterior and posterior touch at day 1 of adulthood. (B) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma and (C) frequency of breaks in ALM and PLM neuronal processes at day 1 of adulthood. (D) Quantification of the incidence of discontinuous ALM neuronal mitochondria. Analysis was done using twnEx8 transgenic animals. (E) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their mechanosensory neurons (takEx415). n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, *p<0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 5—source data 1
Raw data for Figure 5.
- https://doi.org/10.7554/eLife.33052.015
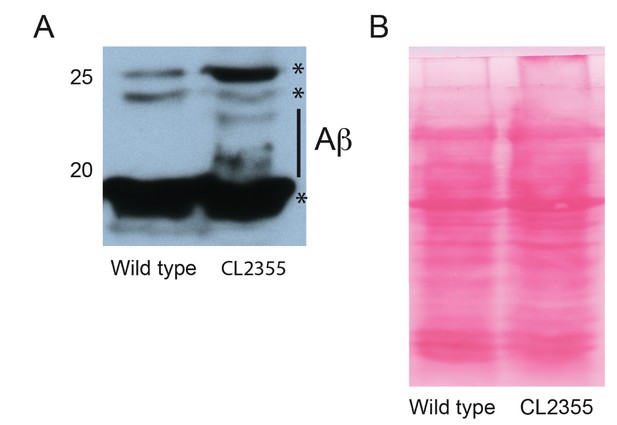
Western analysis of pan-neuronal expression of Abeta1-42 in C. elegans.
(A) Representative western blot indicating Abeta1-42 peptide accumulation in the pan-neuronal Abeta1-42 expressing strain CL2355. * indicate non specific bands as observed in wild type animals. (B) Ponceau C staining of western blot shown in (A) indicating equal loading.
-
Figure 5—figure supplement 1—source data 1
Raw data for Figure 5—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.016
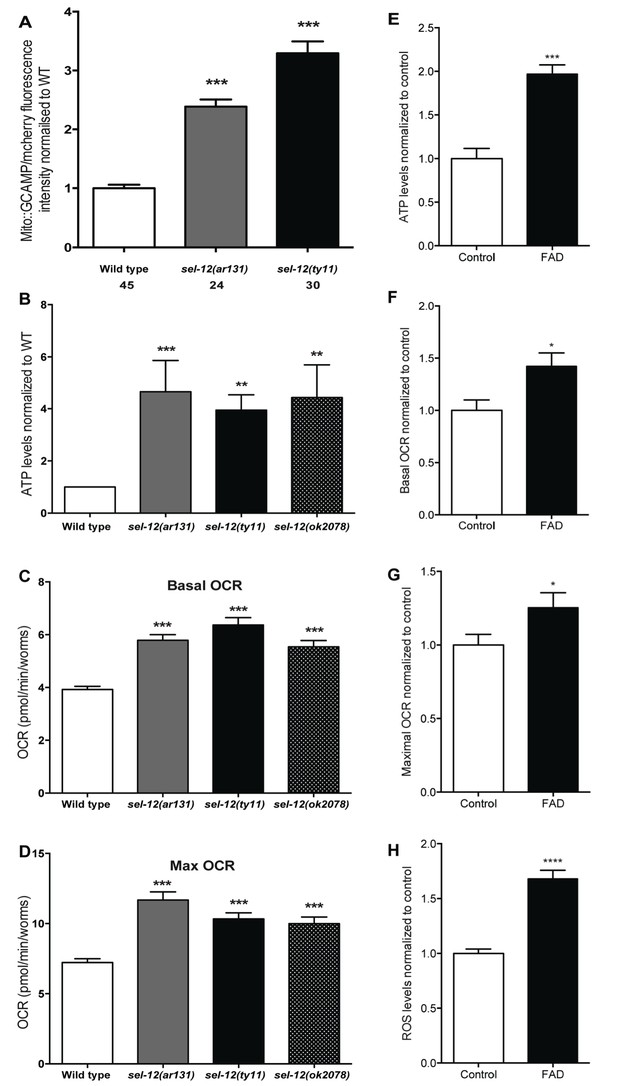
Mutations in sel-12 and PSEN1 result in higher rates of oxygen consumption and generate higher levels of ATP.
(A) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their body wall muscle (takEx347). (B) Quantification of the relative ATP levels in sel-12 mutants compared to wild type animals. Data are from three replicate assays. (C) Basal respiration rates of wild type and sel-12 mutant animals. (D) Maximal respiration rates of wild type and sel-12 mutant animals after exposure to FCCP. OCR data are from three replicate assays. (E) ATP levels normalized to protein content in skin fibroblasts isolated from control and FAD patients. (F) Basal and (G) Maximal OCR normalized to protein content in skin fibroblasts isolated from control and FAD patients. (H). ROS levels normalized to protein content in skin fibroblasts isolated from control and FAD patients. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. *p<0.05, **p<0.01, ***p<0.0001, ****p<0.00001 (One way. ANOVA with Tukey test for A-D, two-tailed T-test for E-H).
-
Figure 1—source data 1
Raw data for Figure 1.
- https://doi.org/10.7554/eLife.33052.019
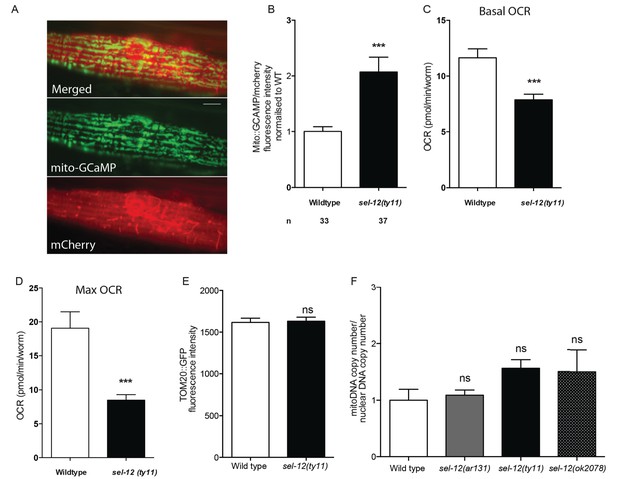
Mitochondrial Ca2+levels are higher in sel-12 mutants.
(A) Representative image of mito-GCaMP6 and mCherry expression (takEx347). Scale bar represents 10 μm. (B) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their mechanosensory neurons (takEx415). (C) Basal and (D) maximal respiration rates of wild type and sel-12 mutant animals at day 8 of adulthood. (E) Mitochondrial fluorescence intensity measured in wild type and sel-12(ty11) animals expressing TOM20::GFP (zcIs14) (F). Quantification of mitochondrial DNA copy number normalized to nuclear DNA copy number in wild type and sel-12(ar131), sel-12(ty11) and sel-12(ok2078) animals. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals. ns p>0.05, ***p<0.0001 (Two-tailed T-test for B-E, one way ANOVA with Tukey test for F).
-
Figure 1—figure supplement 1—source data 1
Raw data for Figure 1—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.020
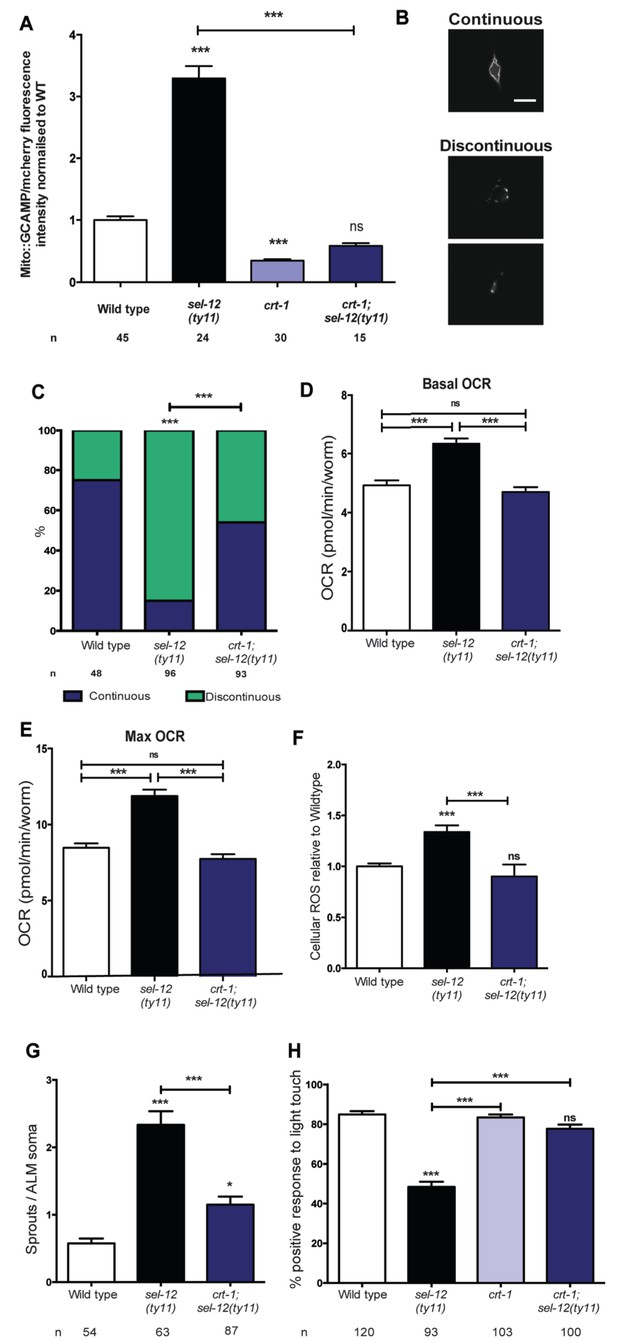
Reducing ER Ca2+release in sel-12 mutants rescues mitochondrial disorganization and dysfunction, ectopic neurite sprouting and mechanosensory defects.
(A) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their body wall muscle (takEx347). n = 30 per strain. (B) Representative images and (C) quantification of ALM neuronal mitochondrial morphology. Scale bar represents 10 μm. Analysis was done using transgenic animals expressing mito::GFP in mechanosensory neurons (jsIs609). Data are displayed with blue and green representing the percentage of animals displaying continuous and discontinuous mitochondrial morphology, respectively. ***p<0.001 (Chi-square test). (D) Basal and (E) maximal respiration rates (after exposure to FCCP) of wild type, sel-12(ty11), and crt-1;sel-12(ty11) animals. Data are from three replicate assays. (F) H2DCF-DA assay measuring ROS levels relative to wild type. Data are from three replicate assays. (G) Quantification of aberrant neuronal structures (sprouts/branches) in ALM neurons in day one animals. ALM neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). (H) Quantification of the response of day one adult animals to light anterior and posterior touch. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, *p<0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 6—source data 1
Raw data for Figure 6.
- https://doi.org/10.7554/eLife.33052.024
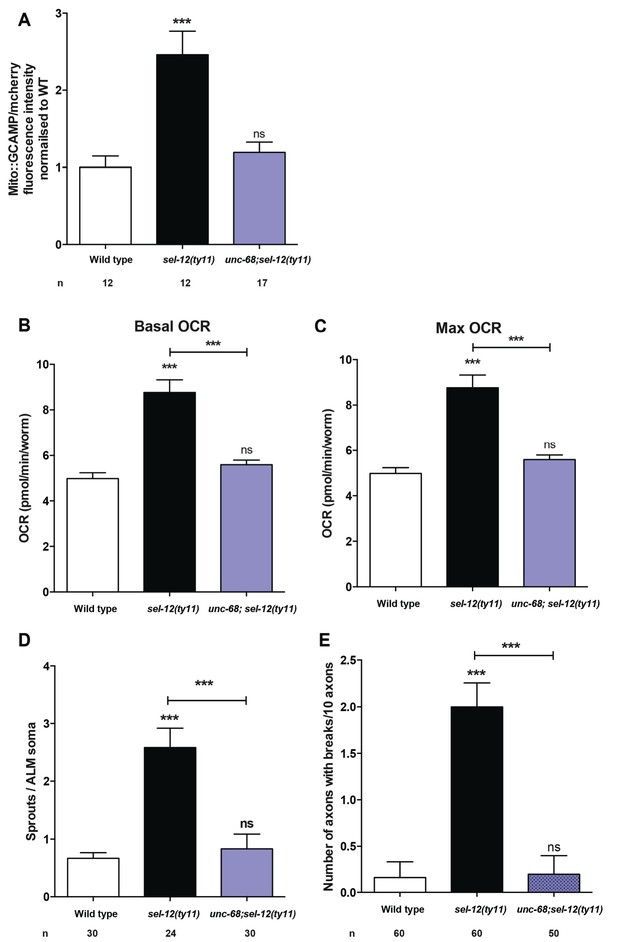
Loss of ryanodine receptors results in the improvement of neurodegeneration in sel-12 null animals.
(A) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their mechanosensory neurons (takEx415). (B) Basal and (C) maximal respiration rates of wild type, sel-12, and unc-68; sel-12(ty11) mutant animals at day 1 of adulthood. (D) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma and (E) frequency of breaks in ALM and PLM neuronal processes at day 1 of adulthood. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 6—figure supplement 1—source data 1
Raw data for Figure 6—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.025
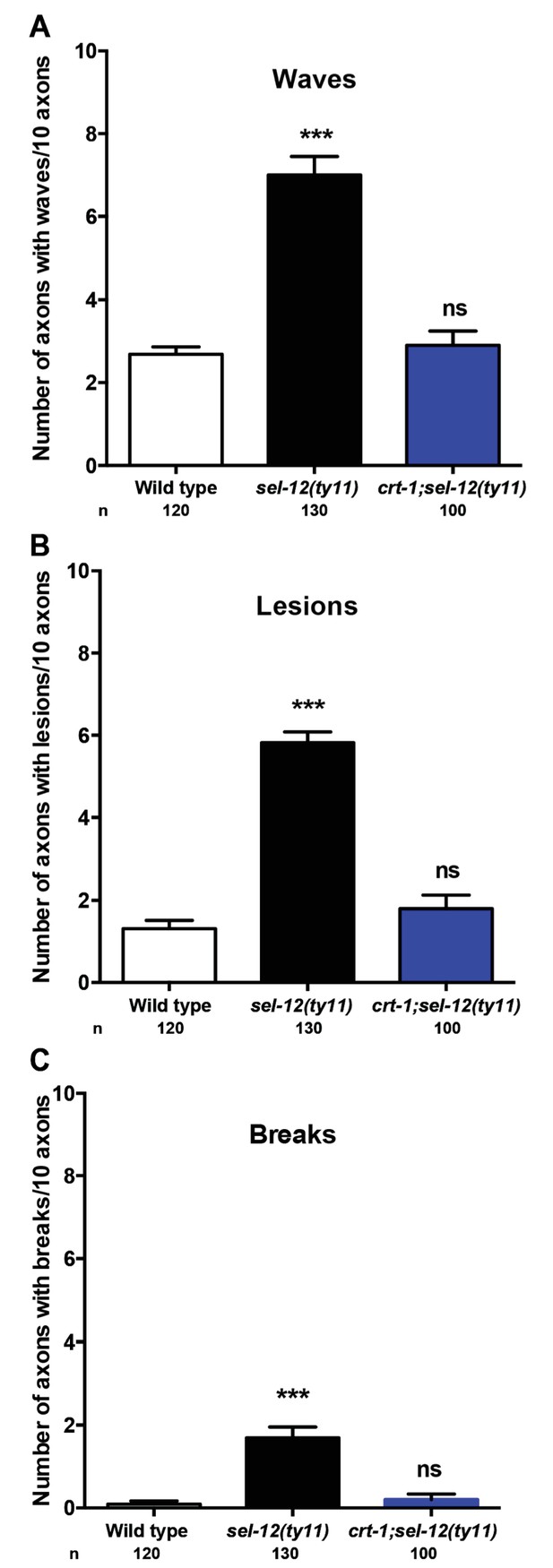
Reduction of ER Ca2+ release improves the structure of mechanosensory neurons in sel-12 animals.
Quantification of the frequency of (A) wave-like processes, (B) lesions and (C) breaks in ALM and PLM neuronal processes in wild type, sel-12(ty11) and crt-1;sel-12(ty11) animals at day 1 of adulthood. Neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 6—figure supplement 2—source data 2
Raw data for Figure 6—figure supplement 2.
- https://doi.org/10.7554/eLife.33052.026
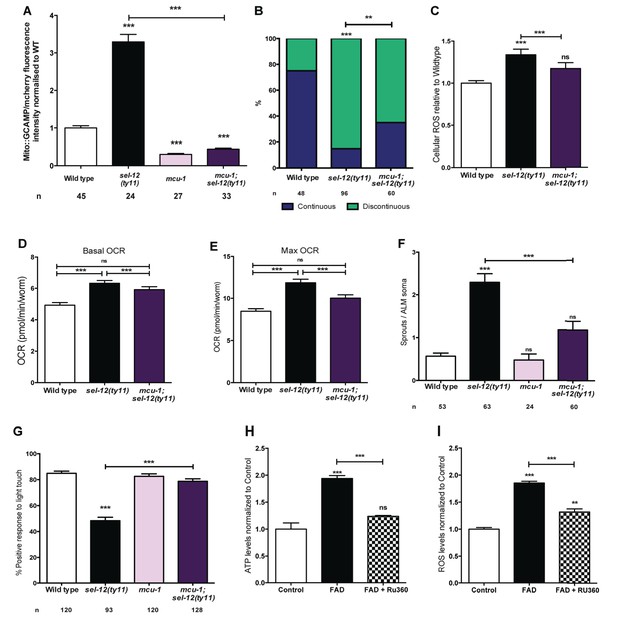
PSEN functions to regulate mitochondrial activity by mediating Ca2+ transfer from the ER to the mitochondria.
(A) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their body wall muscle (takEx347). (B) Quantification of ALM neuronal mitochondrial morphology. Analysis was done using transgenic animals expressing mito::GFP in mechanosensory neurons (jsIs609). Data are displayed with blue and green representing the percentage of animals displaying continuous and discontinuous mitochondrial morphology, respectively. **p<0.01, ***p<0.001 (Chi-square test). (C) H2DCF-DA assay measuring ROS levels relative to wild type. Data are from three replicate assays. (D) Basal and (E) maximal respiration rates (after exposure to FCCP) of wild type, sel-12(ty11), and mcu-1;sel-12(ty11) animals. Data are from three replicate assays. (F) Quantification of aberrant neuronal structures (sprouts/branches) in ALM neurons in day one animals. ALM neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). (G) Quantification of the response of day one wild type, sel-12(ty11), mcu-1;sel-12(ty11) and mcu-1 animals to light anterior and posterior touch. (H) ATP and (I) ROS levels normalized to protein content in skin fibroblasts isolated from control and FAD patients, treated with Ru360. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated). ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey test).
-
Figure 7—source data 1
Raw data for Figure 7.
- https://doi.org/10.7554/eLife.33052.029
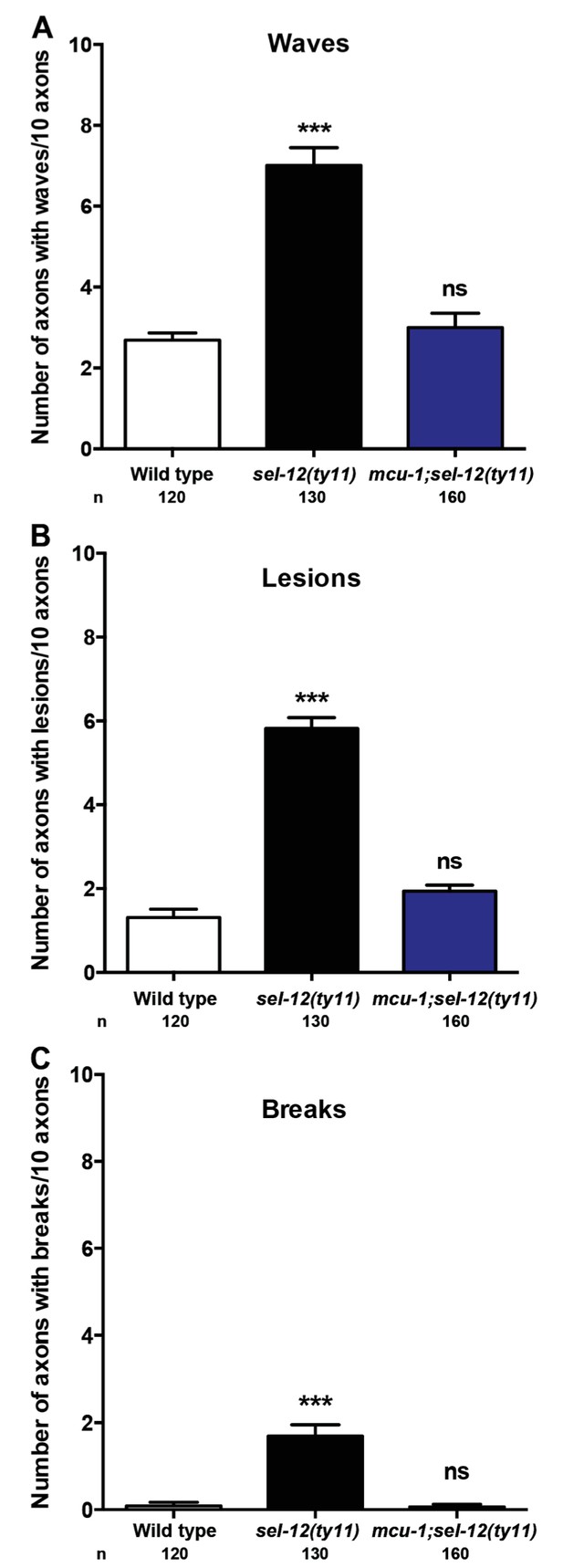
Reduction of ER Ca2+ release improves the structure of mechanosensory neurons in sel-12 animals.
Quantification of (A) wavelike processes, (B) lesions and (C) breaks in ALM and PLM neuronal processes in wild type, sel-12(ty11) and mcu-1;sel-12(ty11) animals at day 1 of adulthood. Neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, ***p<0.0001 (One way ANOVA with Tukey).
-
Figure 7—figure supplement 1—source data 1
Raw data for Figure 7—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.030
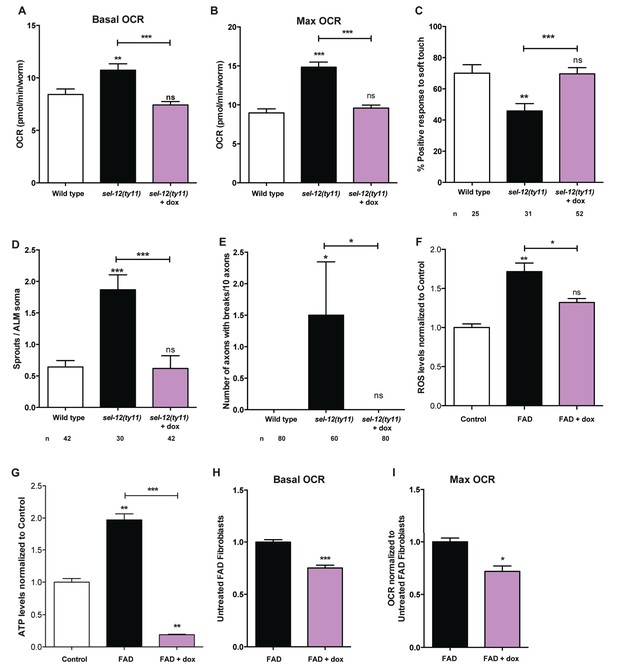
Reducing mitochondrial activity using doxycyline alleviates neurodegenerative phenotypes observed in sel-12 mutants.
(A) Basal and (B) maximal respiration rates of wild type and sel-12 mutant animals treated with doxycycline (dox). (C) Quantification of the response of dox treated day one sel-12(ty11) animals to light anterior and posterior touch. (D) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma and (E) frequency of breaks in ALM and PLM neuronal processes at day 1 of adulthood in response to dox. (F) ROS and (G) ATP levels normalized to protein content in skin fibroblasts isolated from control and FAD patients treated with dox. (H) Basal and (B) maximal respiration rates in skin fibroblasts isolated from control and FAD patients treated with dox. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, *p<0.05, **p<0.001, ***p<0.0001 (One way ANOVA with Tukey test for A-G, two-tailed T-test for H and I).
-
Figure 8—source data 1
Raw data for Figure 8.
- https://doi.org/10.7554/eLife.33052.033
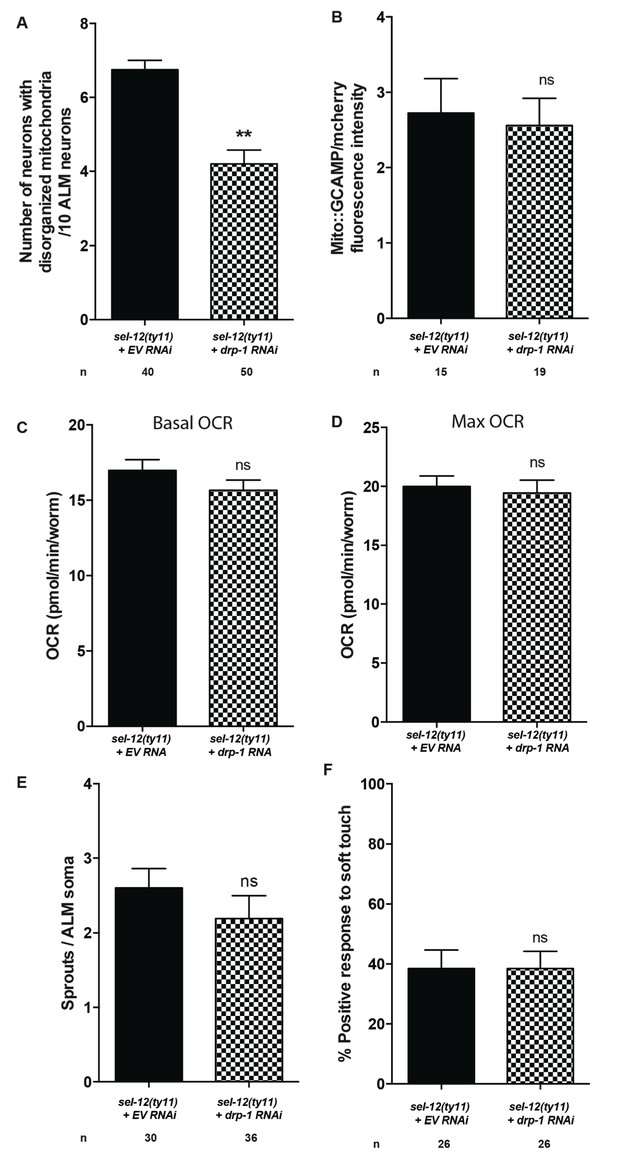
Reduction of mitochondrial fission does not alleviate the neurodegeneration in sel-12(ty11) animals.
(A) Quantification of ALM neuronal mitochondrial morphology in sel-12(ty11) animals grown on EV and drp-1(RNAi). Analysis was done using transgenic animals expressing mito::GFP in mechanosensory neurons (jsIs609). (B) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their mechanosensory neurons (takEx415). (C) Basal and (D) maximal respiration rates of sel-12 mutant animals grown on EV and drp-1(RNAi). (E) Quantification of aberrant structures (sprouts/branches) on ALM neuronal soma of sel-12(ty11) animals grown on EV and drp-1(RNAi). (F) Response of day one sel-12(ty11) animals grown on EV and drp-1(RNAi) to light touch. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated. ns p>0.05, **p<0.001 (two-tailed T-test).
-
Figure 8—figure supplement 1—source data 1
Raw data for Figure 8—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.034
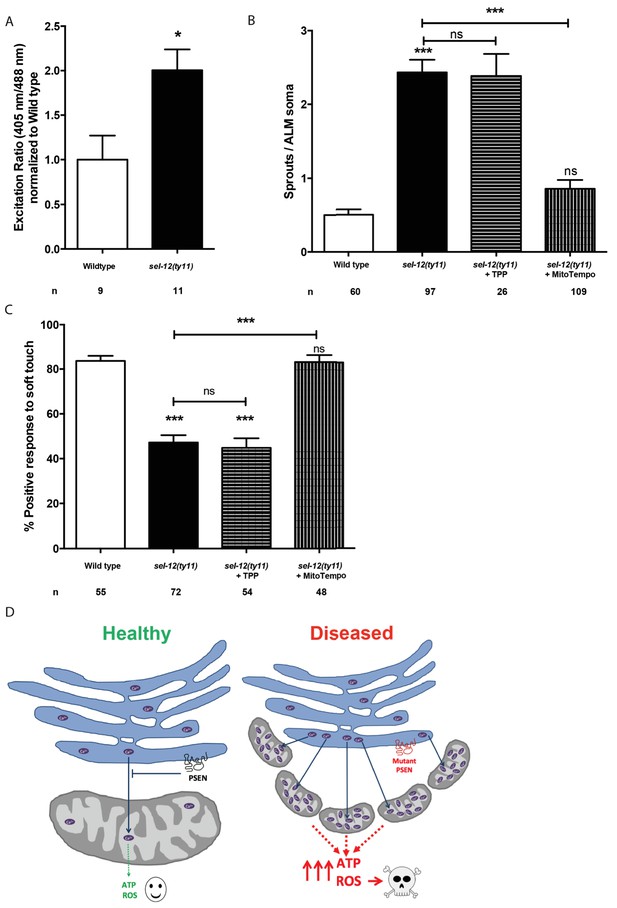
MitoTEMPO supplementation improves neuronal structure and function of sel-12 mutants.
(A) Quantification of oxidized state of ALM neuronal mitochondria using zhsEx17 [Pmec-4mitoLS::ROGFP] (B) Quantification of aberrant neuronal structures (sprouts/branches) in ALM neurons in day one animals. ALM neuronal morphology analysis is done using transgenic animals expressing mec-4p::GFP (zdIs5). (C) Quantification of the response of day one animals to light anterior and posterior touch. n = number of animals analyzed per genotype. Data are displayed as mean ± SEM, and all comparisons have been made to wild type animals unless otherwise indicated). ns p>0.05, *p<0.05, ***p<0.0001 (One way ANOVA with Tukey test and two-tailed t-test for A). (D) Model: Wild type presenilin regulates the normal transfer of Ca2+ (purple ovals) from the ER to the mitochondria. When presenilin is mutated or absent, excessive ER-mitochondria Ca2+ transfer results in mitochondrial morphological changes, increased mitochondrial respiration and ROS production causing neurodegeneration.
-
Figure 9—source data 1
Raw data for Figure 9.
- https://doi.org/10.7554/eLife.33052.037
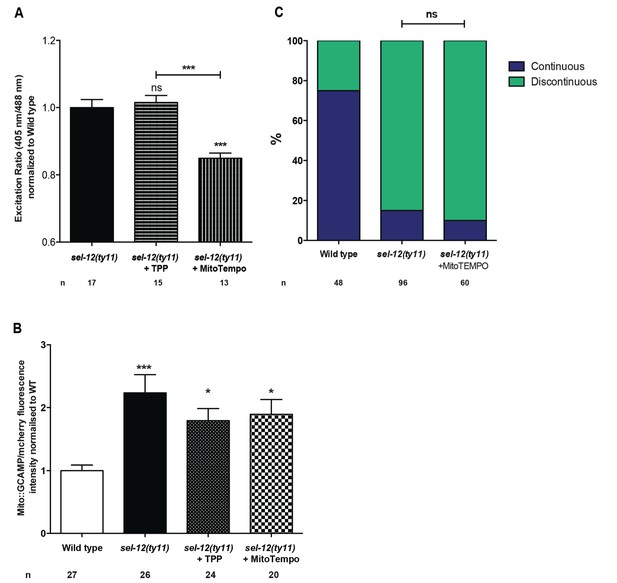
Mitochondrial disorganization in the mechanosensory neurons of sel-12 animals is not a direct result of elevated ROS levels.
(A) Quantification of oxidized state of ALM neuronal mitochondria using zhsEx17 [Pmec-4mitoLS::ROGFP] treated with TPP or mitoTEMPO. (B) Quantification of mitochondrial Ca2+ using animals expressing mito::GCaMP6 and mCherry in their mechanosensory neurons (takEx415) treated with TPP or mitoTEMPO. (C) Quantification of ALM neuronal mitochondrial morphology in wild type and MitoTEMPO or vehicle treated day one sel-12(ty11) animals. Data are displayed with blue and green representing the percentage of animals displaying continuous and discontinuous mitochondrial morphology, respectively. ns p>0.05 (Chi-square test). n = number of animals analyzed per genotype. Mitochondrial analysis was done using transgenic animals expressing mito::GFP in mechanosensory neurons (jsIs609).
-
Figure 9—figure supplement 1—source data 1
Raw data for Figure 9—figure supplement 1.
- https://doi.org/10.7554/eLife.33052.038
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33052.039





