Time- and polarity-dependent proteomic changes associated with homeostatic scaling at central synapses
Figures
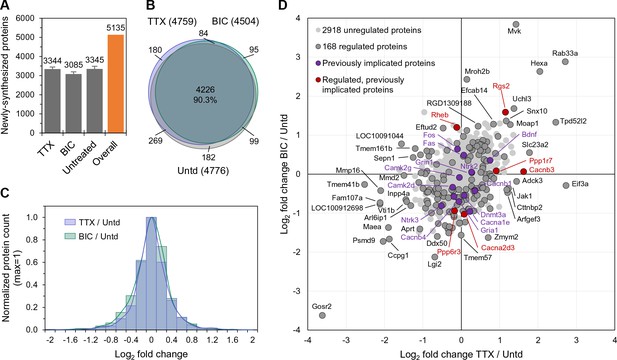
The newly synthesized proteome after 2 hr of enhanced or reduced activity.
(A) Bar chart showing the number of newly synthesized proteins identified on average, in each group as indicated. Error bars represent ±SEM of 5 biological replicates. (B) Venn diagram indicating the number (in parentheses) and overlap of proteins expressed in each treatment group (TTX or bicuculline) and untreated control. (C) Protein fold changes (Log2) showing the overall proteome regulation each treatment group compared to untreated samples. (D) Scatter plot showing all significantly regulated proteins (dark grey; n = 168; ANOVA, FDR = 0.05) as well as the regulation of proteins previously implicated in homeostatic scaling (red). Proteins shown in light grey or purple are not significantly regulated.
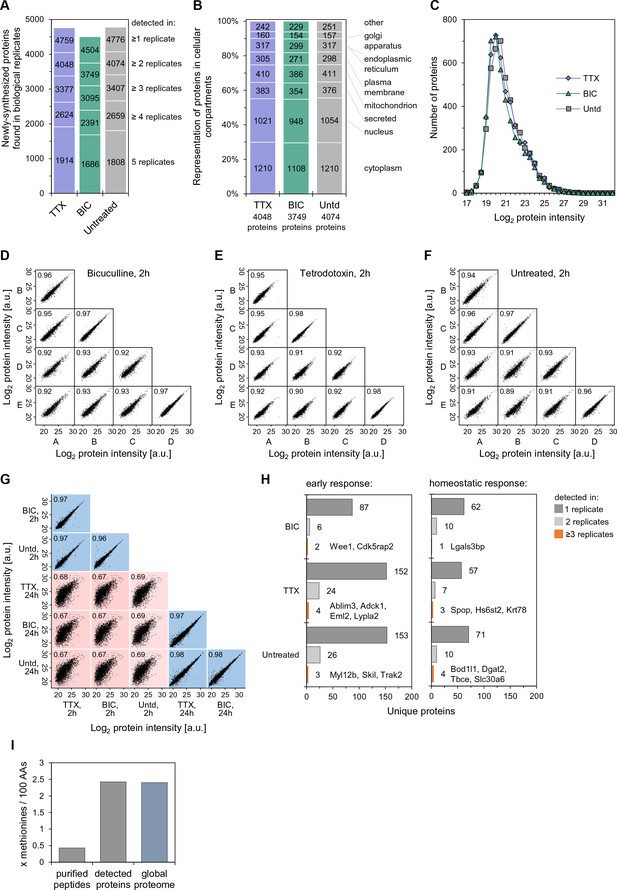
Data reproducibility and overlap of the newly synthesized proteome (2 hr).
(A) Bar chart representing the reproducibility of proteins identified in several biological replicates. Proteins identified in only one replicate were discarded and not analyzed further. (B) Newly-synthesized proteins in each treatment group were classified according to their localization in cellular compartments using Gene Ontology. (C) Distribution of non-normalized, raw protein intensities showing similar protein expression in all conditions. (D–F) Correlation plots of protein intensities across biological replicates for the 2 hr datasets for each treatment, TTX, bicuculline or untreated (control), respectively. (G) Correlation plots of protein intensities for the data sets acquired in this study (2 hr) as well as the data sets in a previous study (Schanzenbächer et al., 2016) that is further analyzed here. In order to be plotted here, the protein must be identified in the two experiments being compared. (H) The unique proteins identified in a single treatment group, showing the number of replicates in which the protein was identified. (I) The methionine content of the peptides identified in our study, proteins identified in our study and the global proteome (from UniprotKB).
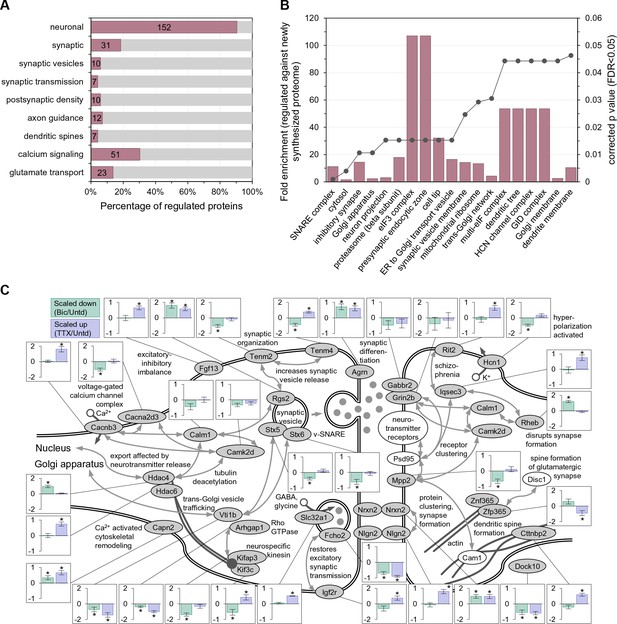
Enriched function groups for proteins that exhibited significant regulation after 2 hr of enhanced or reduced activity.
(A) Gene Ontology enrichment categories for all 168 regulated proteins. A majority of the proteins were associated with the neuronal function group. Note that individual regulated proteins can belong to more than one group. (B) Cellular function enrichment analysis showing the fold-enrichment for the indicated groups (left y-axis, bars) as well as the corrected p value (right y-axis, line). (C) Significantly regulated proteins associated with the synapse and nuclear function. Bar graph inserts show the regulation for scaling-down (bicucculine treatment; pale green bars) or scaling-up (TTX treatment; lavender bars).
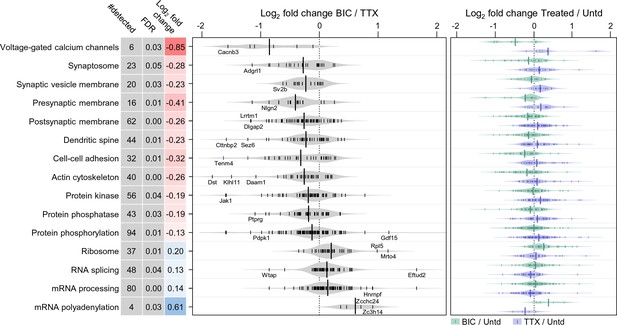
Polarized differential regulation of some protein functional classes after 2 hr of enhanced or reduced activity.
Bean plot of selected significantly regulated protein classes and pathways that exhibited polarized regulation after 2 hr treatment with either TTX or bicucculine. #detected: number of proteins assigned to specified group, FDR: false discovery rate, Log2 fold change: intensity ratio of bicuculline vs TTX treated neurons. Proteins (black markers) were grouped into functional classes and pathways as described in Materials and methods. The bean plots in grey show the comparison between bicucculine and TTX, the green and blue plots show the comparison of each treatment group to a control.
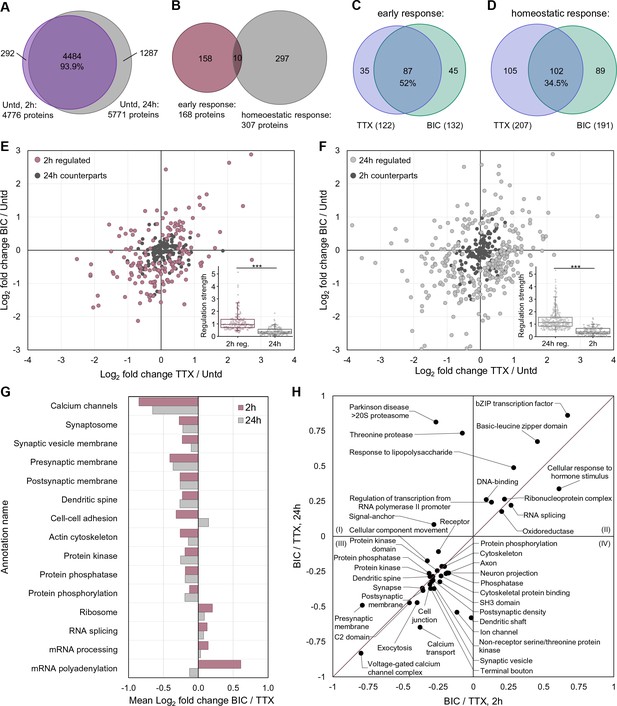
A comparison of proteome remodeling in neurons after 2 hr and 24 hr of synaptic scaling.
(A) Venn diagram showing the overlap of proteins expressed in untreated neurons for 2 hr and 24 hr. (B) Venn diagram showing the overlap of the significantly regulated proteins following 2 hr or 24 hr of synaptic scaling. (C) Venn diagram showing the overlap of the proteins significantly regulated by TTX and bicucculine treatment after 2 hr. The total number of regulated proteins for each group is in parentheses. (D) Venn diagram showing the overlap of the proteins significantly regulated by TTX and bicucculine treatment after 24 hr. The total number of regulated proteins for each group is in parentheses. (E) Scatter plots showing the regulated proteins at 2 hr (mauve dots) and their corresponding regulation at 24 hr (black dots). Inset shows the median regulation strength for the same set of proteins. (F) Scatter plots showing the regulated proteins at 24 hr (grey dots) and their corresponding regulation at 2 hr (black dots). Inset shows the median regulation strength for the same set of proteins. (G) Functional annotation of protein groups showing that, although overlapping individual regulated proteins are rare at 2 and 24 hr, there are similar protein groups that exhibit differential regulation between the up- and down-scaling treatment. (H) More detailed functional characterization of the protein groups that are similarly regulated at 2 and 24 hr.
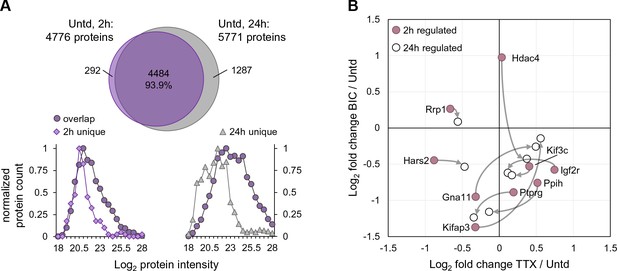
Further analyses of ‘unique’ proteins identified in untreated samples at 2 and 24 hr.
(A) Venn diagram showing the overlap of proteins expressed in untreated neurons for 2 hr and 24 hr together with the Log2 protein intensity for the unique proteins and the overlapping proteins showing that, on average, the unique proteins are less abundant than the overlapping proteins. (B) The Log2 fold change following either TTX or bicucculine treatment for the 10 proteins that we regulated at 2 and 24 hr. The regulation at 2 hr is shown in mauve and the regulation at 24 hr is shown in white.
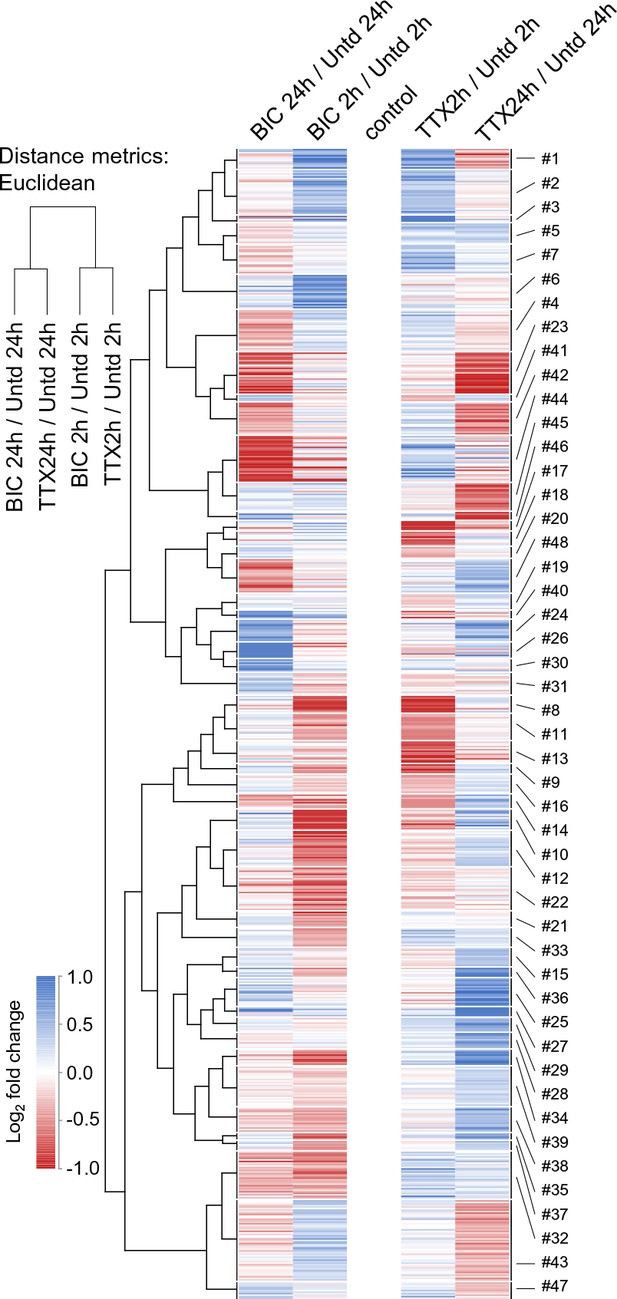
Time-dependent protein regulation in synaptic scaling: patterns of time- and polarity-dependent regulation.
Heat-map showing the clustered patterns of time- and polarity-dependent regulation, for each type of treatment (TTX or bicucculine) and for both (2 and 24 hr) timepoints. The self-organizing maps corresponding to these clusters are shown in Figure 6.
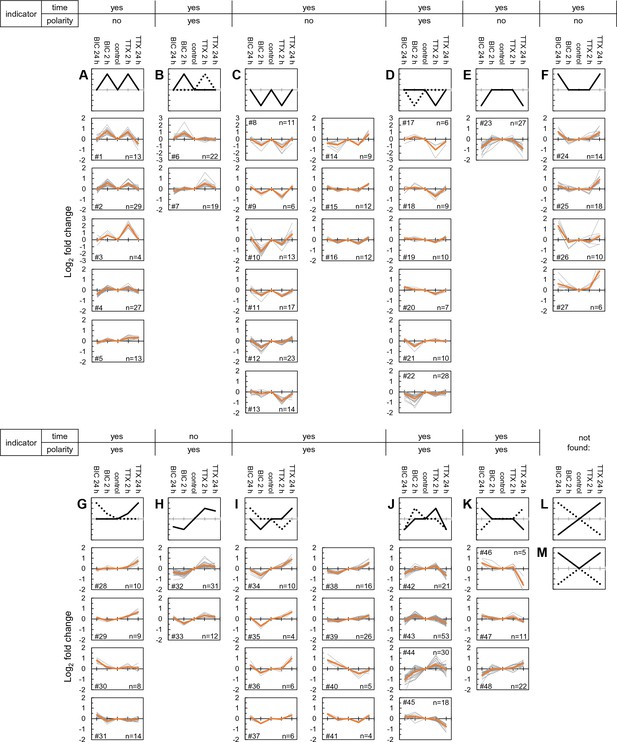
Self-organizing maps define clusters of time- and polarity-dependent proteome regulation.
(A–M) Shown are clusters of regulated protein expression profiles obtained using self-organizing maps. Along the top of the clusters, whether or not the expression profile indicates the time the system has been manipulated (e.g. 2 or 24 hr) or the type of manipulation (up- or down-scaling). (A) The ‘M’ profile, represented by five clusters and a total of 86 different proteins. (B) The modified ‘M’ profile, represented by 2 clusters and 41 different proteins. (C) The ‘W’ profile, represented by nine clusters and a total of 117 different proteins. (D) The modified ‘W’ profile, represented by 6 clusters and 70 different proteins. (E) The ‘inverted trapezoid’ profile represented by 1 cluster of 27 proteins. (F) The ‘trapezoid’ profile represented by 4 clusters and 48 proteins. (G) The ‘sun-seeking worm’ profile represented by 4 clusters and 41 proteins. (H) The ‘sine wave’ profile represented by 2 clusters and 43 proteins. (I) The ‘skewed W’ profile represented by 8 clusters and 77 proteins. (J) The ‘skewed M’ profile represented by 4 clusters and 122 proteins. (K) The ‘flattened trapezoid’ profile represented by 3 clusters and 38 proteins. (L and M) Two profiles that we did not identify in our population of newly synthesized proteins include the ‘diagonal lines’ or the ‘regular or inverted V’.
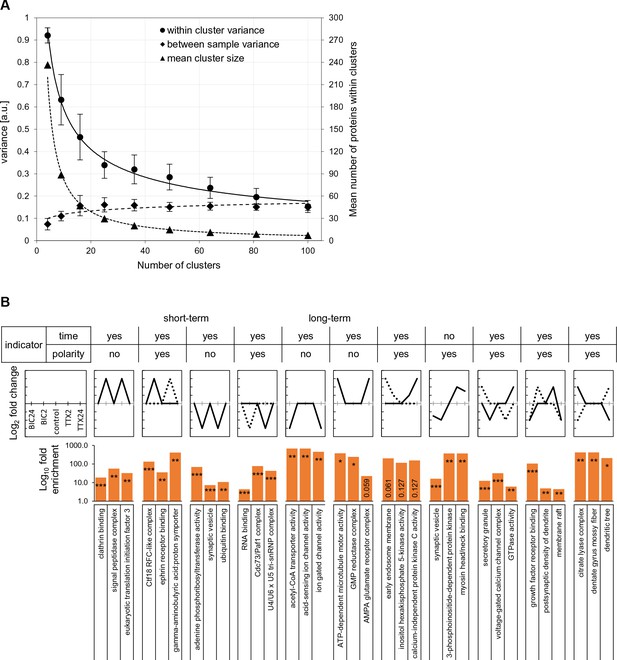
Optimization of cluster size for self-organizing maps and gene ontology analysis of cluster patterns.
(A) In order to determine the optimal cluster size (reducing within-cluster variance and maximizing between-cluster variance) we varied the number of clusters used in the self-organizing maps, also noting the number of proteins per cluster. Based on the data shown in the graph, we chose a cluster size of 48. (B) Gene ontology analysis of significantly enriched protein groups for the various clusters shown in Figure 6.
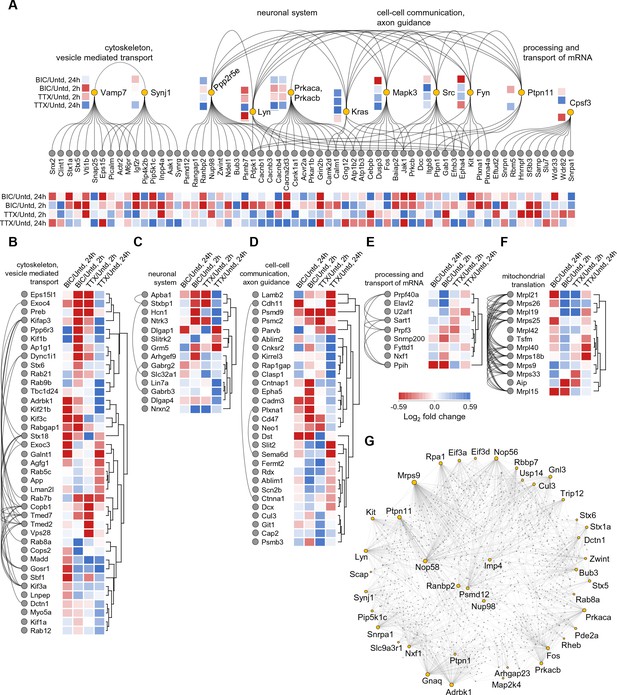
Network analysis of time- and polarity-dependent proteome regulation.
(A–G) Using the String database, we analyzed the patterns of regulation and interaction between regulated proteins, using all 711 regulated proteins. (A) Here we indicate by functional groups, the most interactive proteins and protein groups including (B) cytoskeleton and vesicle-mediated transport, (C) neuronal system, (D) cell-cell communication and axon guidance, (E) processing and transport of RNA, and the highly reciprocally interactive group of regulated proteins (F) associated with mitochondrial translation. (G) The regulated proteins that function as highly interactive hubs in the proteome are depicted.
Additional files
-
Supplementary file 1
Complete lists of newly-synthesized proteins at 2 h and 24 h.
- https://doi.org/10.7554/eLife.33322.013
-
Supplementary file 2
ANOVA of polarity-dependent protein regulation after 2 hrs and previously implicated proteins.
- https://doi.org/10.7554/eLife.33322.014
-
Supplementary file 3
Protein classification and GO enrichment analysis.
- https://doi.org/10.7554/eLife.33322.015
-
Supplementary file 4
Statistical analysis of differentially regulated functional groups.
- https://doi.org/10.7554/eLife.33322.016
-
Supplementary file 5
Significantly regulated proteins in late response (24 hrs) and protein overlap in both timepoints.
- https://doi.org/10.7554/eLife.33322.017
-
Supplementary file 6
ANOVA of time- and polarity-dependent protein regulation and marker enrichment.
- https://doi.org/10.7554/eLife.33322.018
-
Supplementary file 7
Protein clustering and GO enrichment analysis.
- https://doi.org/10.7554/eLife.33322.019
-
Supplementary file 8
Full LC-MS and data analysis parameters.
- https://doi.org/10.7554/eLife.33322.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.33322.021





