Integration of two RAB5 groups during endosomal transport in plants
Figures
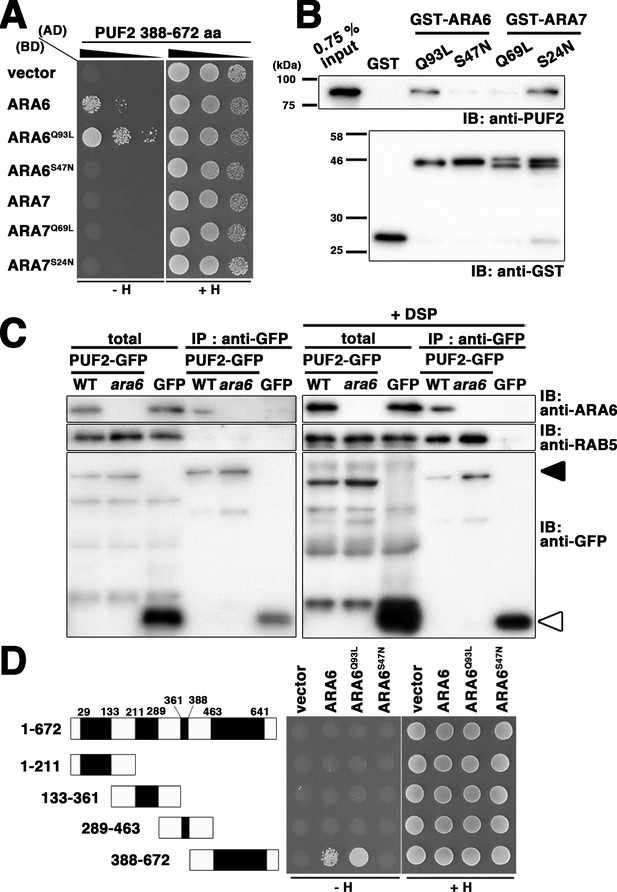
PLANT-UNIQUE RAB5 EFFECTOR 2 (PUF2) isolated as an ARA6 effector.
(A) Yeast two-hybrid interaction between the C-terminal region of PUF2 and RAB5s (ARA6 or ARA7). PUF2 388–672 aa was expressed as a fusion protein with a transcriptional activation domain (AD), and ARA6 and ARA7 with the indicated mutations were expressed as fusion proteins with DNA binding domains (BDs) in the yeast strain AH109. Interactions with the HIS3 reporter gene were examined. Q to L mutations are GTP-freezing mutations, and S to N mutations are GDP-freezing mutations. (B) Interaction between PUF2 and RAB5 members detected in an in vitro binding assay. The full-length PUF2 protein was pulled down using GST-tagged RAB5 proteins, which were immobilized in GTP- or GDP-bound states. (C) Wild-type or ara6-1 transgenic plants expressing PUF2-GFP (black arrowhead) or free GFP (white arrowhead) were subjected to immunoprecipitation analyses with or without a chemical crosslinker (DSP) using an anti-GFP antibody, followed by immunoblotting with the indicated antibodies. A mixture of anti-RHA1 and anti-ARA7 antibodies was used to detect canonical RAB5. (D) Yeast two-hybrid interaction between truncated PUF2 proteins containing different coiled-coil regions and ARA6. Black boxes indicate coiled-coil regions.
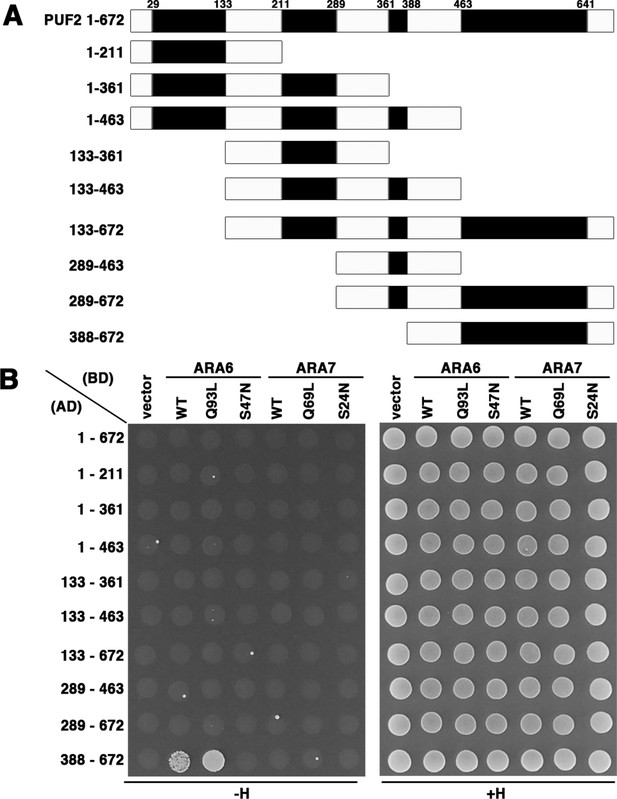
Related to Figure 1.
(A) Schematic diagram depicting the domain structure of PUF2 and the truncated PUF2 constructs used for the assay. Black boxes represent coiled-coil domains. (B) PUF2 constructs in (A) were expressed as fusion proteins with the AD, and RAB5s (ARA6 and ARA7) were expressed as fusion proteins with the BD in the yeast strain AH109. Interactions were tested using the HIS3 reporter gene. Q93L and Q69L are GTP-fixed mutants and S47N and S24N are GDP-fixed mutants of ARA6 and ARA7, respectively.
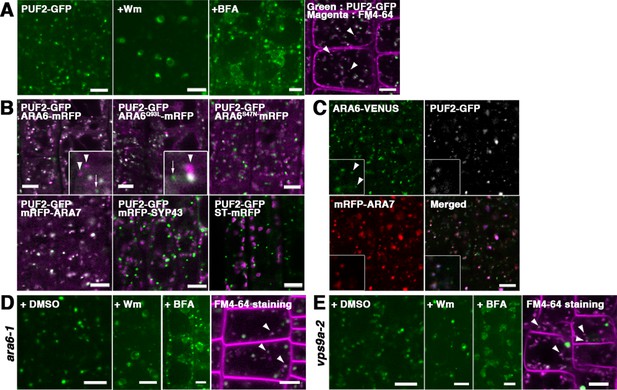
PUF2 preferentially colocalizes with ARA7.
(A) Subcellular localization of PUF2-GFP in root epidermal cells (far-left panel). PUF2-localizing compartments were sensitive to the PI3K inhibitor Wm and an ARF GEF inhibitor, BFA. The PUF2-bearing compartments (green) were accessible to the endocytic tracer FM4-64 (magenta) (far-right panel). Bars = 5 µm. (B) Localization pattern of PUF2-GFP in relation to mRFP-tagged ARA6, ARA6Q93L, ARA6S47N, ARA7, SYP43 (trans-Golgi network marker), and ST (trans-Golgi marker). Puncta bearing only PUF2 and ARA6/ARA6Q93L are indicated with arrows and arrowheads, respectively. Bars = 5 µm. (C) Localization of PUF2-GFP (gray or blue), ARA6-Venus (green), and mRFP-ARA7 (red) in transgenic plants expressing all three tagged proteins. Arrowheads indicate endosomes bearing only ARA6. Bars = 5 µm. (D and E) Responses of PUF2-positive endocytic compartments to Wm or BFA in the ara6-1 (D) and vps9a-2 (E) mutants. PUF2-GFP (green) localized to endocytic compartments stained with FM4-64 (magenta) in these mutants (far-right panels). Bars = 5 µm.
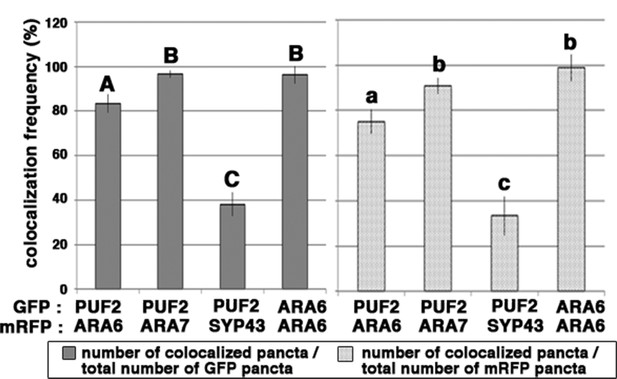
Related to Figure 2.
Bar graph representing the colocalization frequencies of PUF2 and RAB5s. Transgenic lines expressing ARA6-GFP and ARA6-mRFP were analyzed as positive controls. The frequency is presented as the percentage of the number of colocalized dots over the total number of GFP or mRFP dots. Different letters in each column indicate significant difference at p<0.05 by Tukey’s test. Error bars indicate ±the standard deviation.
-
Figure 2—figure supplement 1—source data 1
Quantification of colocalization.
- https://doi.org/10.7554/eLife.34064.007
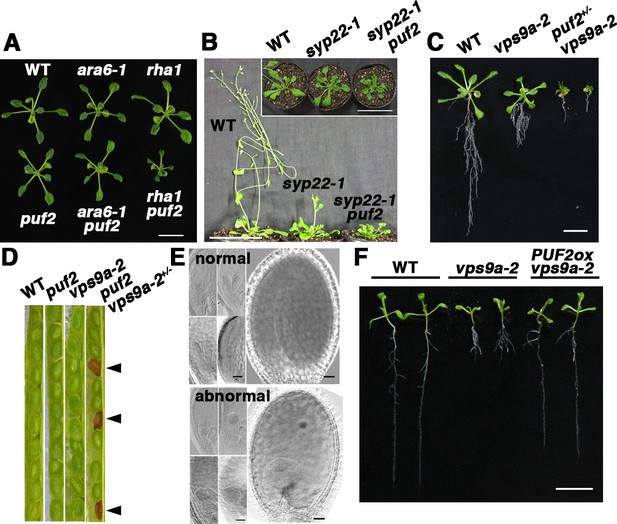
Genetic interaction between PUF2 and endocytic components.
(A) Genetic interactions between PUF2 and ARA6 or RHA1. Fourteen-day-old plants are shown. Bar = 1 cm. (B) Genetic interaction between PUF2 and SYP22. The puf2 mutation aggravated the phenotype of the syp22-1 mutant. The top view of the plants is shown in the inset. Thirty-five-day-old plants are shown. Bars = 5 cm. (C) Genetic interaction between PUF2 and VPS9a. Hemizygous mutation of puf2 (puf2+/−) aggravated the vps9a-2 phenotype. Sixteen-day-old seedlings are shown. Bar = 1 cm. (D) Seed pods collected from wild-type, puf2, vps9a-2 and puf2 vps9a-2+/− plants. Approximately a quarter of the seeds collected from the puf2 vps9a-2+/− plants exhibited a brown and deflated appearance (arrowheads). (E) Embryogenesis of the puf2 vps9a-2 double mutant was delayed and terminated at the globular stage. Corresponding images in the upper and lower sets represent normal and abnormal embryos collected from the same seedpods at different developmental stages. Bars = 50 µm. (F) Overexpression of PUF2 rescued defective primary root elongation of the vps9a-2 mutant. Ten-day-old seedlings are presented. Bar = 1 cm.
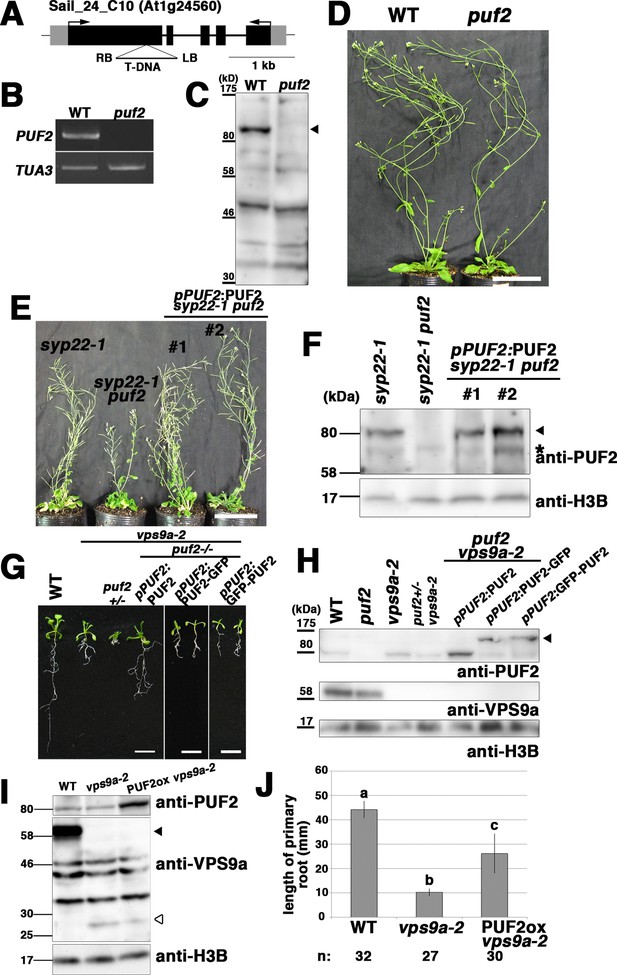
Related to Figure 3.
(A) Schematic structure of the PUF2 gene and position of the T-DNA insertion. LB; left border, RB; right border. (B) RT-PCR analysis of PUF2 expression in 10-day-old seedlings. Expression of the full-length PUF2 CDS was not detected in puf2. Expression of TUA3 was examined as an internal control. WT; wild-type. (C) Immunoblot of plant lysates prepared from 10-day-old wild-type (WT) and puf2 seedlings using an anti-PUF2 antibody. The arrowhead indicates the PUF2 protein. (D) Phenotype of the puf2 single mutant. Thirty-five-day-old plants are shown. Bar = 5 cm. (E) Complementation of the puf2 mutation with a genomic fragment of PUF2. The puf2 syp22-1 mutant plant was transformed. Sixty-two-day-old plants are shown. Bar = 5 cm. (F) Expression of PUF2 (arrowhead) was confirmed by immunoblotting analysis using an anti-PUF2 antibody. Expression of histone 3B (H3B) was examined as an internal loading control. Asterisk indicates the non-specific band. (G) A genomic fragment containing PUF2 was introduced into the puf2 vps9a-2+/− mutant. The genotypes of T1 plants were analyzed by performing PCR-based genotyping to select transgenic lines with the puf2 vps9a-2 double mutation. The lethal phenotype of puf2 vps9a-2 was rescued by PUF2 (left panel) or GFP-tagged PUF2 expressed through the PUF2 promoter (middle and right panels). Fourteen-day-old seedlings are shown. Bars = 1 cm. (H) Expression of the PUF2 product was confirmed by performing immunoblotting analysis using an anti-PUF2 antibody. Expression of H3B was examined as an internal loading control. The arrowhead indicates GFP-tagged PUF2. (I) PUF2 expression levels in the plants shown in Figure 3F were examined by performing immunoblotting analysis. Expression of H3B was examined as an internal loading control. The black and white arrowheads indicate full-length and truncated VPS9a, respectively. (J) Quantification of primary root length presented in Figure 3F. Error bars indicate ±SE.
-
Figure 3—figure supplement 1—source data 1
Quantification of primary root length.
- https://doi.org/10.7554/eLife.34064.010
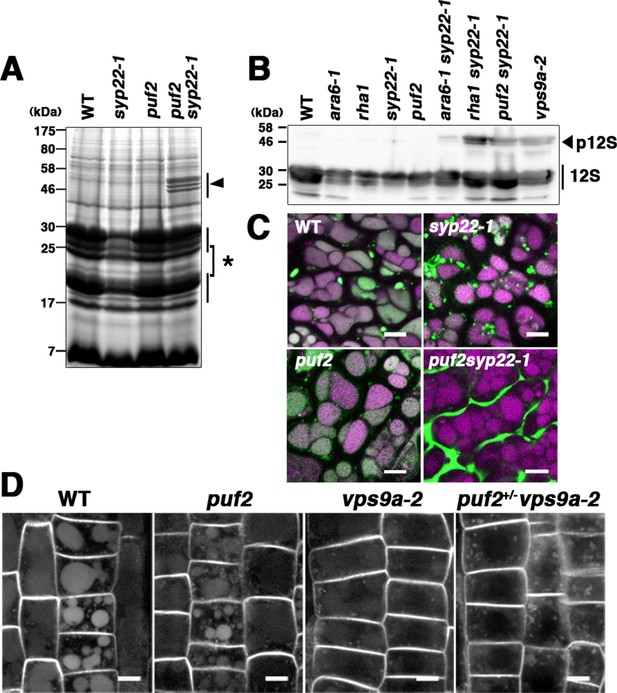
PUF2 is involved in vacuolar trafficking of cargo proteins.
(A) CBB staining of total seed proteins. Unprocessed precursors of storage proteins with large molecular masses were detected in puf2 syp22-1 seeds (arrowhead). The asterisk indicates bands for processed storage proteins. (B) Immunoblotting detection of 12S globulin in seed proteins. Precursors of 12S globulin (p12S, arrowhead) accumulated in puf2 syp22-1 seeds. (C) Distribution of SP-GFP-CT24 in wild-type, syp22-1, puf2, and puf2 syp22-1 embryonic cells. SP-GFP-CT24 (green) was mis-secreted into the extracellular space in the puf2 syp22-1 double mutant. Autofluorescence from protein storage vacuoles is presented in magenta. Bars = 5 µm. (D) Distribution of PIN2-GFP in wild-type, puf2, vps9a-2 and puf2+/− vps9a-2 root cells. Plants were grown vertically under dark conditions for 48 hr prior to observation. Bars = 5 µm.
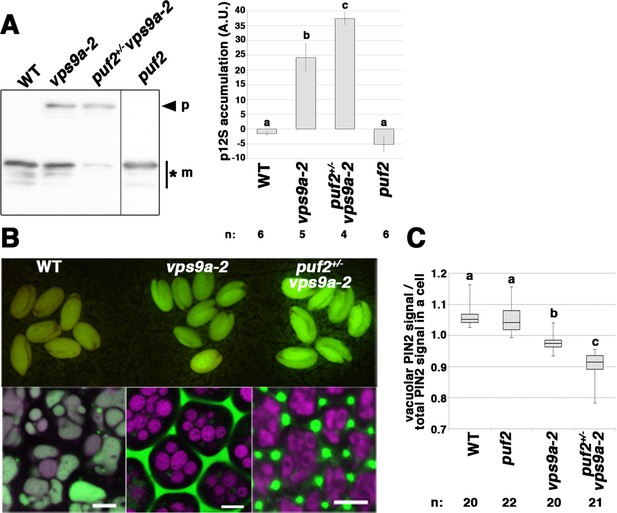
Related to Figure 4.
(A) Immunoblotting analysis of seed proteins collected from WT, vps9a-2, puf2+/− vps9a-2, and puf2 using an anti-12S globulin antibody. High levels of the precursors of 12S globulin (arrowhead) accumulated in puf2+/− vps9a-2 plants. The asterisk indicates bands for processed 12S globulin. The graph on right shows the quantification of the ratio of 12S globulin signal precursors (p) to the total 12S globulin signal (p+m). Error bars indicate ± the standard deviation. (B) puf2+/− vps9a-2 seeds expressing SP-GFP-CT24 exhibited brighter fluorescence compared to vps9a-2 seeds expressing the same cargo protein (upper panel). SP-GFP-CT24 accumulated in the extracellular space in the embryos of vps9a-2 and puf2+/− vps9a-2 mutants (lower panels). Autofluorescence from protein storage vacuoles is shown in magenta. Bars = 5 µm. (C) Quantification of vacuolar accumulation of PIN2-GFP in wild-type (WT), puf2, vps9a-2, and puf2+/− vps9a-2 presented by the box plot. Different letters in each column indicate significant difference at p<0.05 by Tukey’s test.
-
Figure 4—figure supplement 1—source data 1
Quantification of p12S globulin signal.
- https://doi.org/10.7554/eLife.34064.013
-
Figure 4—figure supplement 1—source data 2
Quantification of vacuolar PIN2 signal.
- https://doi.org/10.7554/eLife.34064.014
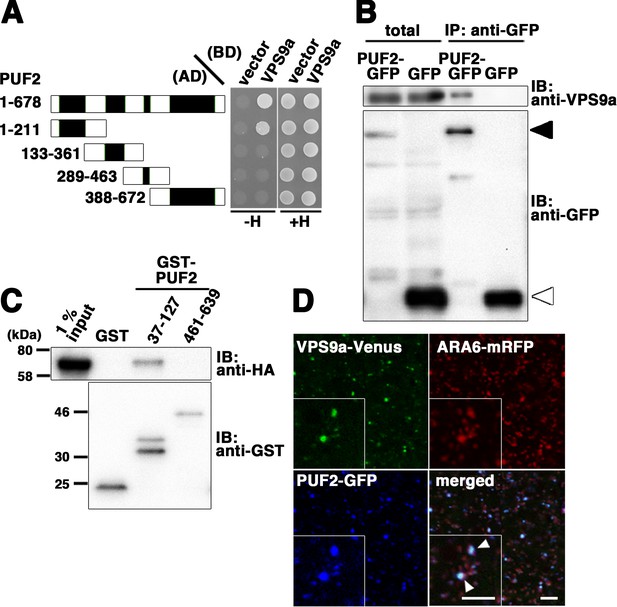
Interaction between PUF2 and the RAB5 activating factor VPS9a.
(A) Yeast two-hybrid interaction between PUF2 and VPS9a. PUF2 containing each coiled-coil region was expressed as a fusion protein with the AD, and VPS9a was expressed as a fusion with the BD in the yeast strain AH109. Interactions were tested using the HIS3 reporter gene. Black boxes indicate coiled-coil regions. (B) Plant lysates prepared from transgenic plants expressing PUF2-GFP (black arrowhead) or free GFP (white arrowhead) were subjected to immunoprecipitation with an anti-GFP antibody, followed by immunoblotting using the indicated antibodies. (C) The interaction between PUF2 and VPS9a was detected using an in vitro binding assay. Yeast lysate containing VPS9a-HA was subjected to a pull-down assay with GST-fused truncated PUF2 protein. VPS9a-HA interacted with PUF2 37–127 aa containing the N-terminal coiled-coil region but not with PUF2 461–639 aa containing the C-terminal coiled-coil region. VPS9a-HA did not interact with GST. Loading was performed with 1% input. (D) Localization of VPS9a-Venus (green), ARA6-mRFP (red), and PUF2-GFP (blue) expressed in the same plant. VPS9a and PUF2 colocalized on a subpopulation of ARA6-positive endosomes. Bars = 5 µm.
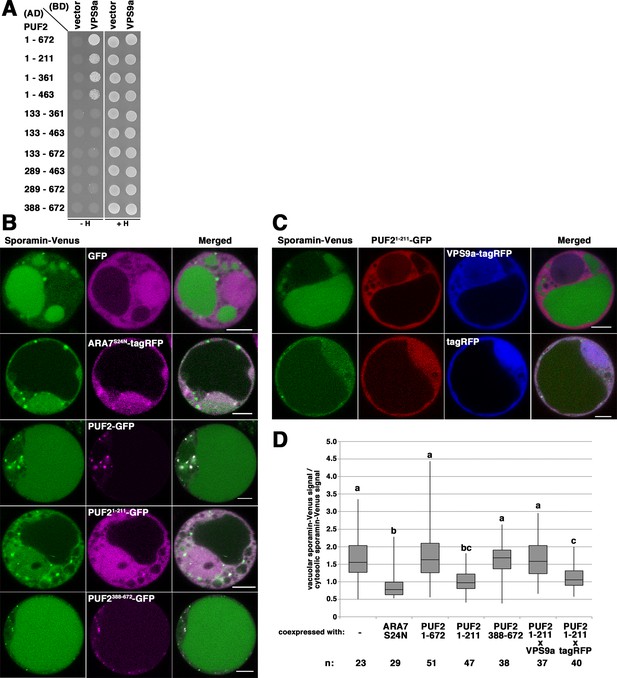
Related to Figure 5.
(A) The PUF2 constructs shown in Figure 1—figure supplement 1A were expressed as fusion proteins with the AD, and VPS9a was expressed as the fusion protein with the BD in the yeast strain AH109. Their interaction was tested using the HIS3 reporter gene. (B) Effects of N-terminal (PUF21-211-GFP) and C-terminal (PUF2388-672-GFP) coiled-coil regions of PUF2 on vacuolar transport of sporamin-Venus (green) in protoplasts prepared from Arabidopsis suspension cultured cells. Vacuolar signal from sporamin-Venus (green) was reduced when co-expressed with tagRFP-ARA7S24N or PUF21-211-GFP (magenta), whereas co-expression of GFP, full-length PUF2 tagged with GFP (PUF2-GFP), or PUF2388-678-GFP (magenta) did not markedly affect the vacuolar transport of sporamin. Bars = 5 µm. (C) The defective vacuolar transport of sporamin-Venus (green) caused by PUF21-211-GFP (red) was rescued by co-expression of VPS9a-tagRFP (blue, upper panels), but not by tagRFP alone (blue, lower panels). Bars = 5 µm. (D) Quantification of vacuolar sporamin-Venus signals presented by box plot. Protoplasts with the vacuole occupying 50% to 75% of the cell volume were analyzed. Different letters in each column indicate significant difference at p<0.05 by Tukey’s test.
-
Figure 5—figure supplement 1—source data 1
Quantification of vacuolar Sporamin-Venus signal.
- https://doi.org/10.7554/eLife.34064.017
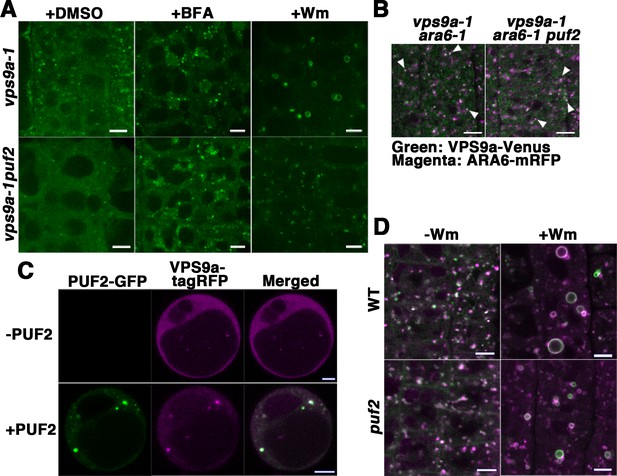
PUF2 is required for the efficient recruitment of VPS9a onto endosomes as well as endosomal fusion.
(A) Localization of VPS9a-GFP in the root epidermal cells of vps9a-1 and vps9a-1 puf2 plants treated with DMSO, BFA, or Wm. Bars = 5 µm. (B) Localization of VPS9a-Venus (green) and ARA6-mRFP (magenta) in vps9a-1 ara6-1 and vps9a-1 ara6-1 puf2 cells. Arrowheads indicate endosomes bearing both VPS9a-GFP and ARA6-mRFP. Bars = 5 µm. (C) Overexpression of PUF2-GFP recruits VPS9a onto punctate compartments in protoplasts. VPS9a-tagRFP (magenta) was localized to punctate structures following the overexpression of PUF2-GFP (green) and dispersed throughout the cytosol in the absence of PUF2-GFP. Bars = 5 µm. (D) Localization of ARA6-GFP (green) and mRFP-ARA7 (magenta) in the root epidermal cells of the puf2 mutant treated with DMSO (−Wm) or Wm (+Wm). Bars = 5 µm.
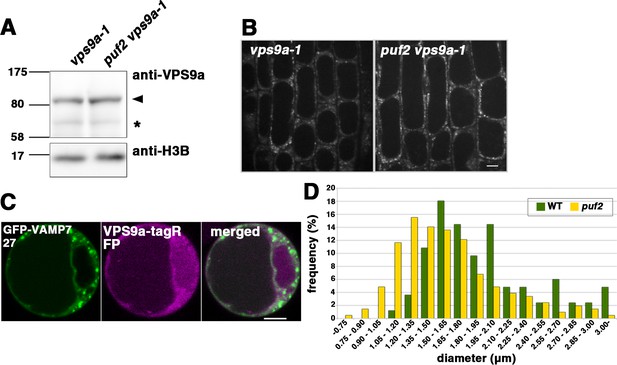
Related to Figure 6.
(A) The expression levels of VPS9a-GFP in vps9a-1 and puf2 vps9a-1 were comparable. The expression of H3B was examined as an internal loading control. The arrowhead indicates VPS9a-GFP, and the asterisk indicates the non-specific band. (B) Distribution of VPS9a-GFP in vps9a-1 and puf2 vps9a-1 lateral root cap cells. Bar = 5 µm. (C) Effects of GFP-VAMP727 (green) overexpression on the localization of VPS9a-tagRFP (magenta) in protoplasts of Arabidopsis suspension-cultured cells. Bar = 5 µm. (D) Histogram depicting the size distribution of dilated ring-shaped structures following Wm treatment in WT (yellow) and puf2 (green) plants. The Feret’s diameters of the structures were measured using ImageJ software.
-
Figure 6—figure supplement 1—source data 1
Wortmannin ring size.
- https://doi.org/10.7554/eLife.34064.020
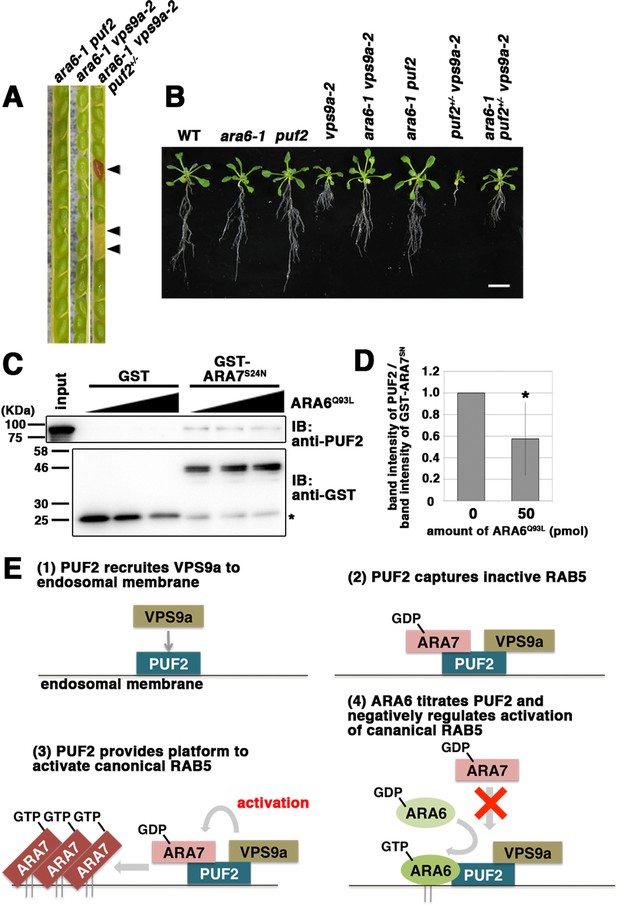
PUF2 integrates functions of two plant RAB5 groups.
(A) Seed pods collected from wild-type, ara6-1 vps9a-2, and ara6-1 vps9a-2 puf2+/− plants. Arrowheads indicate brown and deflated seeds in a seed pod from the ara6-1 vps9a-2 puf2+/− plant. (B) Fourteen-day-old seedlings with the indicated genotypes. Bar = 1 cm. (C) Interaction between ARA7S24N and PUF2 pre-incubated with ARA6Q93L. The assay was performed as described for Figure 1B, but PUF2 was pre-incubated with 0 pmol, 25 pmol or 50 pmol ARA6Q93L before mixing with GST-ARA7S24N. Loading was performed with 1.5% input. (D) Quantification of the band intensities of PUF2, calibrated based on the band intensities of GST-ARA7S24N. The error bar indicates the mean ±SD. *, p<0.05. (F) Proposed model for the integration of two plant RAB5 groups mediated by PUF2. PUF2 recruits VPS9a and the inactive form of canonical RAB5 onto the endosome, leading to the efficient activation of canonical RAB5 and RAB5-dependent trafficking events. GTP-bound active ARA6 negatively regulates this process by competitively binding to the PUF2-VPS9a complex.
-
Figure 7—source data 1
Quantification of PUF2 band intensity.
- https://doi.org/10.7554/eLife.34064.023
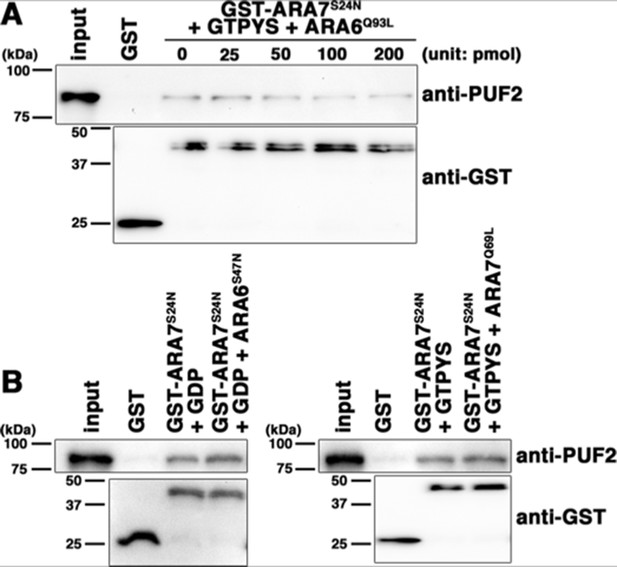
Related to Figure 7.
(A) Dosage-dependent reduction of the interaction between ARA7S24N and PUF2 by ARA6Q93L. PUF2 was pre-incubated with 0, 25, 50, 100, or 200 pmol ARA6Q93L and 10 µM GTPγS before mixing with GST-ARA7S24N. 2% input was also loaded (input). (B) Interaction between ARA7S24N and PUF2 pre-incubated with non-interactive RAB5, ARA6S47N (left) or ARA7Q69L (right). PUF2 was pre-incubated with 0 pmol or 50 pmol non-interactive RAB5 and 10 µM GDP (for ARA6S47) or GTP (for ARA7Q69L) before mixing with GST-ARA7S24N. 2% input was also loaded (input).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Arabidopsis thaliana) | PUF2 | NA | TAIR:At1g24560 | |
| Gene (A. thaliana) | ARA6/RABF1 | NA | TAIR:At3g54840 | |
| Gene (A. thaliana) | RHA1/RABF2a | NA | TAIR:At5g45130 | |
| Gene (A. thaliana) | ARA7/RABF2b | NA | TAIR:At4g19640 | |
| Gene (A. thaliana) | SYP43 | NA | TAIR:At3g05710 | |
| Gene (A. thaliana) | VPS9a | NA | TAIR:At3g19770 | |
| Gene (A. thaliana) | SYP22 | NA | TAIR:At5g46860 | |
| Gene (A. thaliana) | VAMP727 | NA | TAIR:At3g54300 | |
| Cell line (A.thaliana) | Deep | PMID:9681013 | ||
| Genetic reagent (A.thaliana) | puf2 | This paper | TAIR:SAIL_24_C10 | |
| Genetic reagent (A.thaliana) | ara6-1 | PMID:21666683 | TAIR:SAIL_880_C07 | |
| Genetic reagent (A.thaliana) | rha1 | PMID:17468262 | TAIR:SAIL_596_A03 | |
| Genetic reagent (A.thaliana) | syp22-1 | PMID:18984676 | TAIR:SALK_060946 | |
| Genetic reagent (A.thaliana) | vps9a-2 | PMID:18055610 | TAIR:GABI_557C02 | |
| Strain, strain background (Saccharomyces cerevisiae) | AH109 | PMID:8978031 | ||
| Strain, strain back ground (S. cerevisiae) | YPH414 | PMID:18055610 | ||
| Strain, strain background (Agrobacterium tumefaciens) | GV3101::pMP90 | other | widely distributed | |
| Strain, strain background (Escherichia coli) | DH5α | PMID:6345791 | ||
| Strain, strain background (E. coli) | Rosetta-gami DE3 | Merck | Merck:71351-3CN | |
| Antibody | anti-PUF2; anti-MBP-PUF2 | This paper | (1:200) | |
| Antibody | anti-GFP | This paper | (1:1,000) | |
| Antibody | anti-12S globulin | PMID:14657332 | (1:10,000) | |
| Antibody | anti-HA(rabbit polyclonal) | Themo Fisher Scientific | Themo Fisher Scientific:71–5500; RRID:AB_2533988 | (1:500) |
| Antibody | anti-GST(rabbit polyclonal) | Santa Cruz Biotechnology | Santa Cruz Biotechnology:sc-459;RRID:AB_631586 | (1:1,000) |
| Antibody | anti-H3B(rabbit polyclonal) | Merck | Merck: 07–473; RRID:AB_1977252 | (1:1,000) |
| Antibody | anti-ARA6(rabbit polyclonal) | PMID:17468262 | (1:200) | |
| Antibody | anti-ARA7(rabbit polyclonal) | PMID:17468262 | (1:200) | |
| Antibody | anti-RHA1(rabbit polyclonal) | PMID:21666683 | (1:200) | |
| Antibody | anti-VPS9a(rabbit polyclonal) | PMID:18055610 | (1:1,000) | |
| Recombinant DNA reagent | pHGW (vector) | PMID:11992820 | ||
| Recombinant DNA reagent | pGWB (vector) | PMID:17697981 | ||
| Recombinant DNA reagent | pGEX6P-1 (vector) | GE Healthcare | GE Healthcare:28954648 | |
| Recombinant DNA reagent | pTS911 (vector) | PMID:26493488 | ||
| Commercial assay or kit | GAL4 Two-Hybrid Phagemid Vector Kit | Agilent Technologies | Agilent Technologies:211351 | |
| Commercial assay or kit | Protein Fusion and Purification System | NewEngland BioLabs | NewEngland BioLabs:800 | |
| Commercial assay or kit | uMACS GFP isolation kit | Miltenyi Biotec | Miltenyi Biotec:130-091-288 | |
| Chemical compound, drug | DSP | Themo Fisher Scientific | Themo Fisher Scientific:22585 | |
| Software, algorithm | Fiji | PMID:22743772 | RRID:SCR_002285 | |
| Software, algorithm | SMART | PMID:25300481 | RRID:SCR_005026 | |
| Other | Wortmannin | Sigma-Aldrich | Sigma-Aldrich:W1628-1MG | |
| Other | Brefeldin A | Sigma-Aldrich | Sigma-Aldrich:B7651-5MG | |
| Other | FM4-64 | Themo Fisher Scientific | Themo Fisher Scientific:T13320 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34064.024





