Modulation of formin processivity by profilin and mechanical tension
Figures
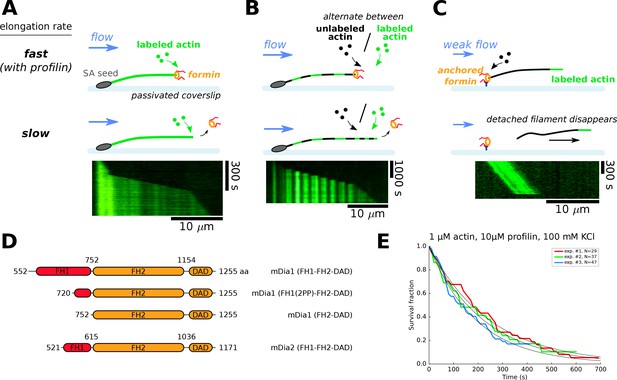
Single-filament microfluidics experimental configurations to measure formin processivity.
(A–C) Different experimental configurations using microfluidics for the study of formin processivity, showing sketches of the side view (top) and typical kymographs of individual filaments (bottom). (A) Alexa 488 labeled actin filaments are elongated from surface-anchored spectrin-actin seeds. Transient exposure to a formin solution puts formins on filament barbed ends, which elongate faster (here in the presence of 1 µM 15% Alexa 488 labeled actin +5 µM profilin, at 100 mM KCl). Upon formin dissociation, the barbed end elongates slower. Images were acquired in TIRF microscopy. See Video 1. (B) Same configuration as in (A), but the filaments are exposed to a periodic alternation of different conditions: here a solution of unlabeled actin (0.3 µM actin, 50 mM KCl) for 100 s and a solution of 15% Alexa 488 labeled actin (0.5 µM actin +2 µM profilin, 50 mM KCl) for 20 s. Images were acquired in epifluorescence while exposing to unlabeled actin. See Video 2. (C) Configuration where formins are anchored to the surface by their C-terminus. Filaments were nucleated using a solution of labeled actin and elongated by flowing in a solution of unlabeled actin (here, 0.3 µM actin, at 50 mM KCl), until the filaments eventually detached and disappeared. The viscous drag applied on the filaments was kept low (<0.1 pN) by working with low flow rates. Images were acquired in epifluorescence. See Video 3. (D) Domain architecture and boundaries for the mDia1 and mDia2 formin constructs used in this study. (E) Survival fractions of mDia1(FH1-FH2-DAD) formin-bound barbed ends as a function of time, obtained from three independent experiments performed in the same conditions, in the experimental configuration shown in (A). Curves are fitted by a mono-exponential decay to obtain formin dissociation rate koff.
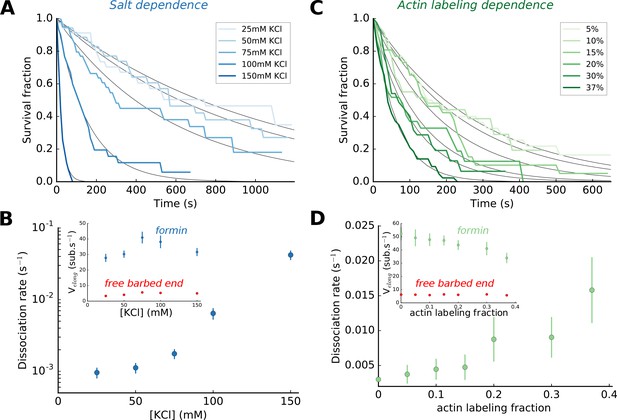
Impact of salt and actin labeling fraction on mDia1 formin processivity.
(A,B) Effect of salt concentration on the survival fraction of formin-bound barbed ends (A), on the formin dissociation rates (B, log-linear scale) as well as on the barbed end elongation rates (B, inset). The dissociation rates in (B) result from the exponential fits (black lines) shown in (A). Each data point corresponds to a population of 30–40 filaments. (C,D) Effect of the actin Alexa 488 labeling fraction on the survival fraction of formin-bound barbed ends (C), on the formin dissociation rates (D) and on the barbed end elongation rates (D, inset). Each data point in (D) corresponds to a population of 30–40 filaments. Error bars on formin dissociation rates indicate 65% confidence intervals based on exponential fits and sample size (see Materials and methods), and error bars on elongation rates indicate standard deviations.
-
Figure 2—source data 1
Spreadsheet containing the data plotted in Figure 2 and Figure 2—figure supplement 1.
Units and conditions are indicated in the figure legends. The data are on separate sheets, as indicated on the thumbnails.
- https://doi.org/10.7554/eLife.34176.008
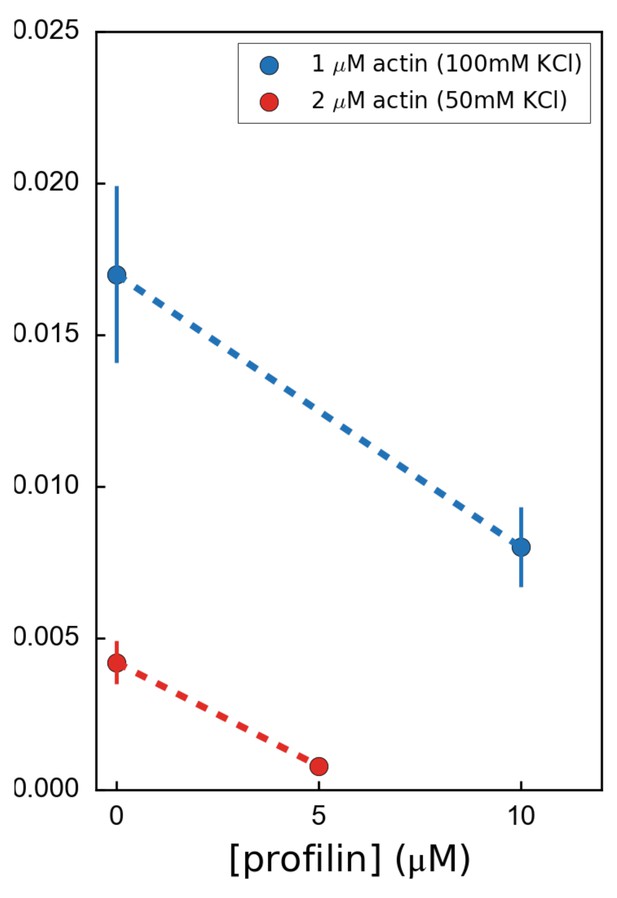
Variation of the formin mDia1(FH1-FH2-DAD) dissociation rate as a function of profilin concentration, for 2 µM unlabeled actin at 50 mM KCl (red) or for 1 µM unlabeled actin at 100 mM KCl (blue), showing that formin processivity is decreased by profilin with unlabeled actin, for both salt conditions.
https://doi.org/10.7554/eLife.34176.007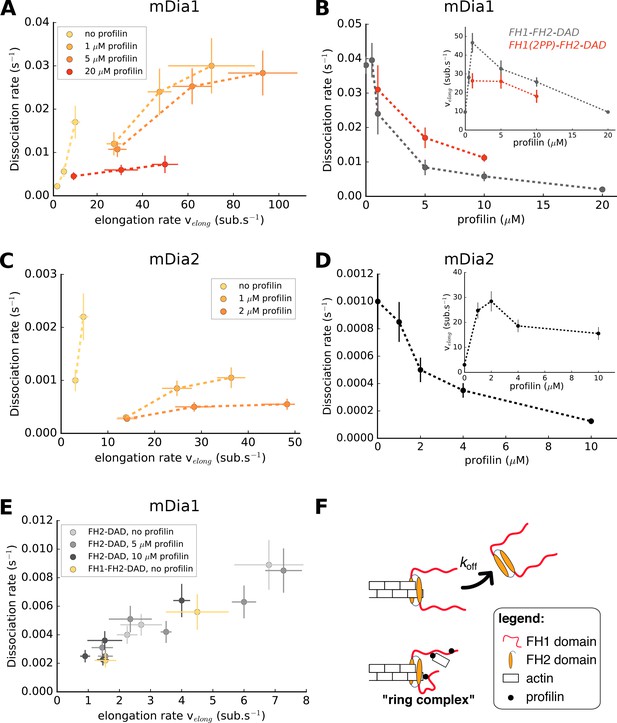
Formin dissociation is enhanced by G-actin concentration, and slowed down by profilin.
(A, C) Variation of the mDia1 (A) or mDia2 (C) formin dissociation rate as a function of the barbed end elongation rate. Each data set (N = 30–40 filaments) was obtained with a fixed profilin concentration and different actin concentrations, at 100 mM KCl. Each point corresponds to an independent experiment, performed with 15% Alexa 488-labeled actin, except for the data of mDia1 without profilin which were acquired with unlabeled actin (as described in Figure 1B). (B, D) Variation of the formin dissociation rate and the barbed end elongation rate (inset) as a function of profilin concentration, (B) for mDia1 formins with a full length FH1 (FH1-FH2-DAD) and with a truncated FH1 containing only two polyproline tracks (FH1(2PP)-FH2-DAD), and (D) for mDia2(FH1-FH2-DAD). The data was obtained with 15% Alexa 488-labeled actin, at 100 mM KCl. The same profilin dependence was observed using unlabeled actin, for both 50 and 100 mM KCl (Figure 2—figure supplement 1). Each data point corresponds to the average of 1–3 independent experiments. (E) Variation of the formin dissociation rate as a function of the barbed end elongation rate: for mDia1(FH2-DAD) homodimers in the presence or absence of profilin, and for mDia1(FH1-FH2-DAD) in the absence of profilin, all with unlabeled actin. (F) Sketch illustrating the profilin-mediated interaction between FH1 and the barbed end, forming the ‘ring complex’, which appears to prevent the dissociation of formin from the barbed end. Error bars on formin dissociation rates indicate 65% confidence intervals based on exponential fits and sample size (see Materials and methods), and error bars on elongation rates indicate standard deviations.
-
Figure 3—source data 1
Spreadsheet containing the data plotted in Figure 3, Figure 3—figure supplement 1, and Figure 3—figure supplement 2.
Units and conditions are indicated in the figures legends. The data are on separate sheets, as indicated on the thumbnails.
- https://doi.org/10.7554/eLife.34176.012
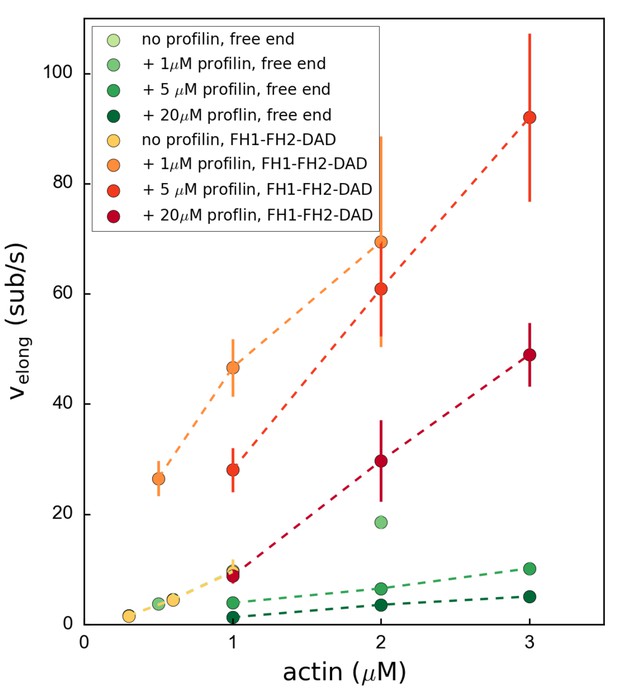
Variation of the mDia1 formin-bound or free barbed end elongation rate velong as a function of actin concentration.
Each data point corresponds to an independent experiment (N = 30–40 filaments) which was conducted at 100 mM KCl. Error bars indicate standard deviations.
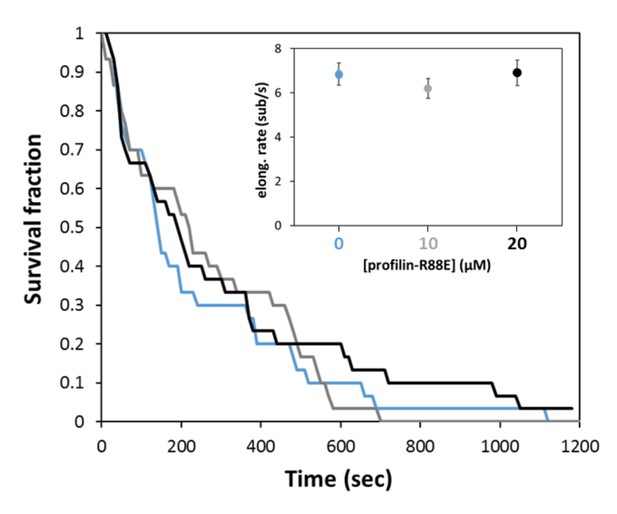
Mutant profilin-R88E has no impact on the processivity of mDia1.
Survival fraction of mDia1 formin-anchored filaments (N = 30 for each curve), exposed to negligible force (<0.1 pN) while elongating with 1 µM actin and either zero (blue), 10 µM (gray) or 20 µM profilin-R88E mutant (black). Inset: average elongation rates measured in the same conditions (subset of N = 11–13 filaments, for each condition). Error bars represent standard deviations.
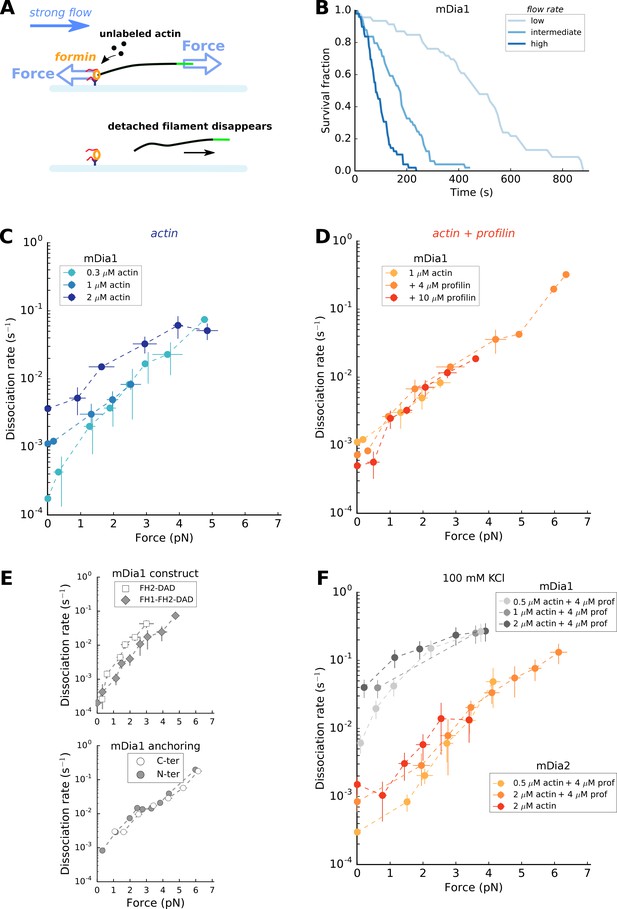
Force has a great impact on formin processivity.
(A) Sketch of the experimental configuration, similar to that of Figure 1C, but where significant forces are applied using various flow rates. The applied force scales with the filament length. (B) Survival fractions of mDia1-anchored filaments, elongating with 1 µM actin +10 µM profilin, using different flow rates to reach different force ranges: each filament underwent 0.051 pN/µm (initial filament length = 4.9 µm, N = 46 filaments), 0.204 pN/µm (initial filament length = 3.2 µm, N = 49) or 0.501 pN/µm (initial filament length = 2.6 µm, N = 49) for ‘low’, ‘medium’ and ‘high’ flow rate curves, respectively. (C–E) mDia1 formin dissociation rate as a function of applied force (log-linear plots), for different actin concentrations in the absence of profilin (C); for 1 µM actin with different profilin concentrations (D); for 0.3 µM actin in presence or absence of FH1 domains (E, top); and for 1 µM actin, 4 µM profilin for mDia1 (FH1-FH2-DAD) formins either anchored by their FH1 N-terminus or FH2 C-terminus (E, bottom). Experiments were carried out by elongating the filaments with unlabeled actin, at 50 mM KCl. (F) mDia1 and mDia2 formin dissociation rates as a function of applied force (log-linear plots), for different profilin and unlabeled actin concentrations, at 100 mM KCl. Dissociation rates were obtained by local fits of the slope in survival fractions similar to the ones shown in (B) (see Materials and methods). Each data point is either obtained from a single experiment or is the average of 2–3 independent experiments. The data points at zero force were measured independently, using the configuration shown in Figure 1B (striped filaments). The error bars indicate standard deviations when several independent experiments were grouped (data from individual experiments for (C) and (D) are shown in Supp. Figure 4—figure supplement 2).
-
Figure 4—source data 1
Spreadsheet containing the data plotted in Figure 4, Figure 4—figure supplement 1, Figure 4—figure supplement 2, and Figure 4—figure supplement 3.
Units and conditions are indicated in the figures legends. The data are on separate sheets, as indicated on the thumbnails.
- https://doi.org/10.7554/eLife.34176.017
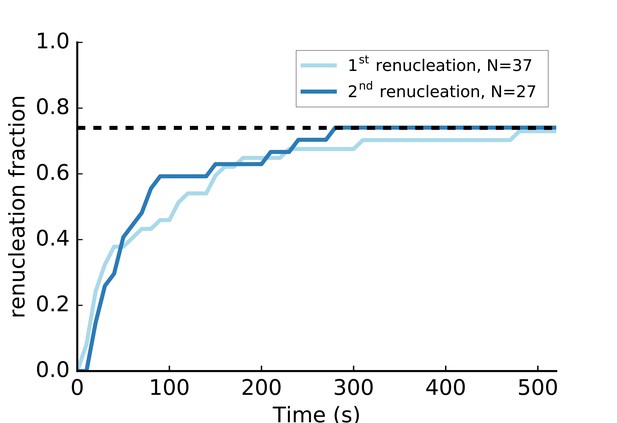
C-terminus anchored mDia1 (FH1-FH2-DAD) formin renucleation.
Nucleation of new filaments from surface-anchored mDia1 formins. The anchored mDia1(FH1-FH2-DAD) formins that participated in a pulling force experiment with 1 µM actin 4 µM profilin in a moderate flow (i.e. reaching a pulling force ~1–4 pN before filaments detached) as depicted in Figure 4, were subjected to a renucleation assay to assess if they were still present on the coverslip surface and functional. The renucleation assay consisted in exposing them alternatively, at the same flow rate, to a solution of F-buffer at 25 mM KCl, 2 µM 15% Alexa 488-labeled actin, 0.4 µM profilin and to a solution of F-buffer at 100 mM KCl, 1 µM unlabeled actin, 4 µM profilin, for 15 s each. The formins that were observed renucleating a filament (‘1 st renucleation’, light blue), eventually let go of that filament and could be observed nucleating another filament (‘2nd renucleation’, dark blue). The dashed line indicates that 74% of the formins had nucleated a new filament after 500 s.
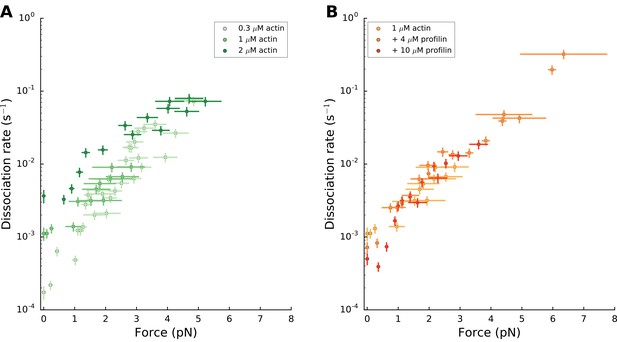
mDia1 formin dissociates faster with force.
mDia1 formin dissociation rate as a function of applied force (log-linear plots): for different actin concentrations in the absence of profilin (left); for 1 µM actin with different profilin concentrations (right). Dissociation rates were obtained by local fits of the slope in survival fractions similar to the ones shown in Figure 4B (see Materials and methods). Each data point is obtained from a single experiment. Error bars indicate the standard deviations for force and the ±0.13% uncertainty on the dissociation rate, accounting for formin detachments from the surface (see Materials and methods). Grouping these data in bins of similar forces resulted in the plots shown in Figure 4C and D.
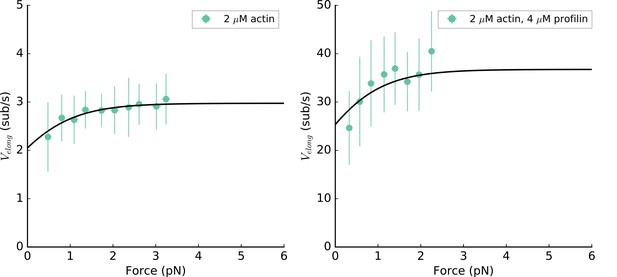
mDia2 formin elongation rate is mechanosensitive.
C-terminus anchored mDia2 (FH1-FH2-DAD) formin elongation rate as a function of the applied tension for 2 µM unlabeled actin in absence (left) or in presence of 4 µM profilin (right), at 100 mM KCl. Data from both conditions are simultaneously fitted, according to the model from (Jégou et al., 2013), resulting in the probability to be in the open state p0 = 69%, and kon = 1.5 and 18.4 µM−1.s−1 in the absence or presence of profilin, respectively.
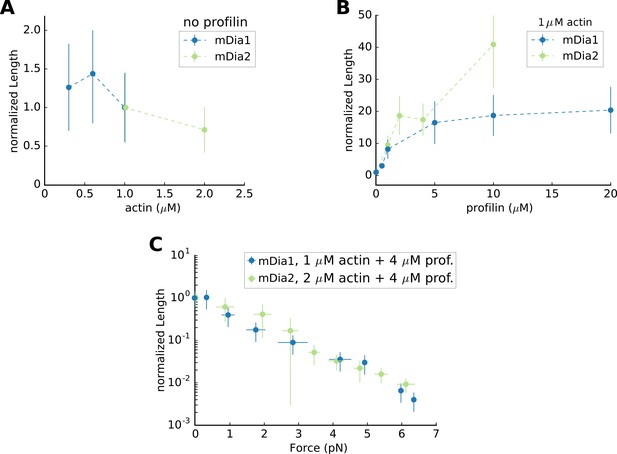
Mean Filament Length generated by mDia1 or mDia2 formins.
Mean flament length (A) as a function of actin concentration, in the absence of profilin, normalized by its value at 1 µM actin (which equals 0.65 µm for mDia1 and 8.22 µm for mDia2); (B) mean length as a function of profilin concentration, at 1 µM actin, normalized by its value in the absence of profilin; (C) mean length as a function of force, at 2 µM actin 4 µM profilin for mDia2, and at 1 µM actin 4 µM profilin for mDia1, normalized by its value in the absence of force (83.7 µm for mDia2 and 7.45 µm for mDia1). All data were acquired at 100 mM KCl. Error bars indicate standard deviations.
-
Figure 5—source data 1
Spreadsheet containing the data plotted in Figure 5.
Units and conditions are indicated in the figures legends. The data are on separate sheets, as indicated on the thumbnails.
- https://doi.org/10.7554/eLife.34176.019
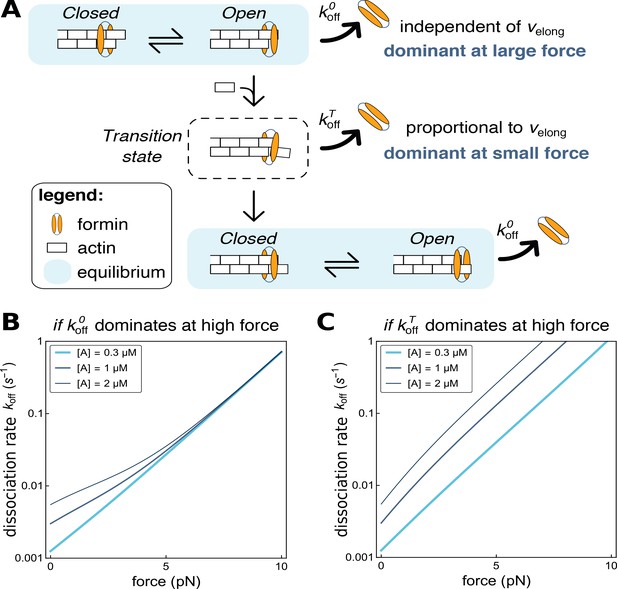
Modeling formin dissociation, in the absence of profilin.
(A) Sketch summarizing the conformations adopted by the FH2 dimer and the actin filament barbed end in our model, in the absence of profilin (for a complete description of the model, see Appendix 1). The system is in rapid equilibrium between an open and a closed state (depicted here as in the ‘stair-stepping’ model) and only the open state allows the addition of a new actin subunit at the barbed end. Following this elongation event, the system is in a transition state, which decays rapidly into a new open-closed rapid equilibrium. Formin dissociation from the barbed end can occur while the system is in the open state (with rate koffO) or in the transition state (rate koffT). The global, observable dissociation rate koff comprises these two routes. (B,C) Predictions of the model for the variation of the dissociation rate koff as a function of force, in log-linear representations. In both cases, koffT is the dominant contribution at zero force. In B, koffO increases more strongly than koffT when force is applied and thus becomes dominant at high force (computed with working distances δO=δ and δT=0, see Appendix 1). In C, koffT increases more strongly than koffO when force is applied (δO=0 and δT=δ, see Appendix 1).
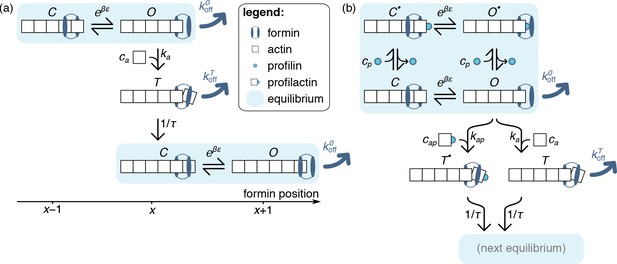
Model of formin function based on transitions between discrete states, as described in the text.
Unlike those of the main text, the schematics presented here only picture one actin protofilament for simplicity, without any implications for the model itself. In both panels, formin dissociation from states and is indicated by thick dark blue arrows. The FH1 domains, which lead to the formation of a ring complex that hampers this dissociation in profilin-associated states, is not explicitely represented here. (a) Simplified, no-profilin model introducing the notion of rapid equilibrium between monomer additions, and transit through a short-lived state upon monomer addition. (b) Model taking into account the association of profilin with the filament barbed end.
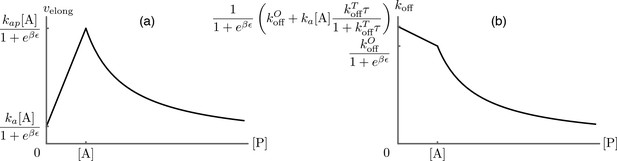
Predictions for the elongation velocity (a) and the formin dissociation rate (b) as functions of the profilin concentration within the approximation of strong actin-profilin binding of Eq.(A13).
While the exact position of the curves is dependent on the choice of parameters as indicated on the figure, their qualitative shapes are a robust prediction of the model, and agree well with the experimental data presented in Figure 3B of main text.
Videos
Actin filaments elongating with 1 µM 15% Alexa 488-labeled actin and 5 µM profilin, at 100 mM KCl, are transiently exposed to a solution of 20 nM mDia1(FH1-FH2-DAD) for 20 s (during frames 27–30).
Images were acquired in TIRF. Full field of view is 137 × 137 µm. Interval between images is 5 s (movie is accelerated 75x). The solution flows from left to right. Corresponds to Figure 1A of the main text.
Actin filaments are exposed to a periodic alternation of a solution of unlabeled actin (0.3 µM actin, 50 mM KCl) for 100 s and a solution of 15% Alexa 488-labeled actin (0.5 µM actin +2 µM profilin, 50 mM KCl) for 20 s.
The filaments are transiently exposed to a solution of 11 nM mDia1(FH1-FH2-DAD) for 5 s, after frame number 5. Images were acquired in epifluorescence while exposing to unlabeled actin. Full field of view is 137 × 137 µm. Interval between images is 120 s (movie is accelerated 360x). The solution flows from left to right. Corresponds to Figure 1B of the main text.
Actin filaments were nucleated from surface-anchored formins mDia1(FH1-FH2-DAD) with 15% Alexa 488-labeled actin, and elongate with 0.3 µM unlabeled actin, at 50 mM KCl.
Full field of view is 221 × 221 µm. Images were acquired in epifluorescence. Interval between images is 10 s (movie is accelerated 70x). A minimal flow is applied. The solution flows from left to right. Corresponds to Figure 1C of the main text.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | BL21(DE3) | Thermo Fischer | Cat# C600003 | |
| Biological sample (Rabbit) | Rabbit muscle | INRA Jouy-en-Josas | N/A | |
| Peptide, recombinant protein | Mouse mDia1(FH1-FH2-DAD) | Uniprot | O08808 | seq. 552–1255 aa |
| Peptide, recombinant protein | Mouse mDia2(FH1-FH2-DAD) | Uniprot | Q9Z207 | seq. 521–1171 aa |
| Peptide, recombinant protein | Human profilin-1 | Uniprot | P07737 | |
| Antibody | PentaHis Biotin conjugate | Qiagen | 34440 | |
| Chemical compound, drug | Alexa Fluor 488 succinimidyl ester | Life Technologies | Cat#A20000 | |
| Commercial assay or kit | Protino Ni-NTA Agarose beads | Macherey-Nagel | Cat#745400.25 | |
| Commercial assay or kit | HiLoad 16/60 Superdex 200 gel filtration column | GE Healthcare | Cat#28-9893-35 | |
| Software, algorithm | numpy/scipy packages | Python |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34176.021





