Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice
Figures
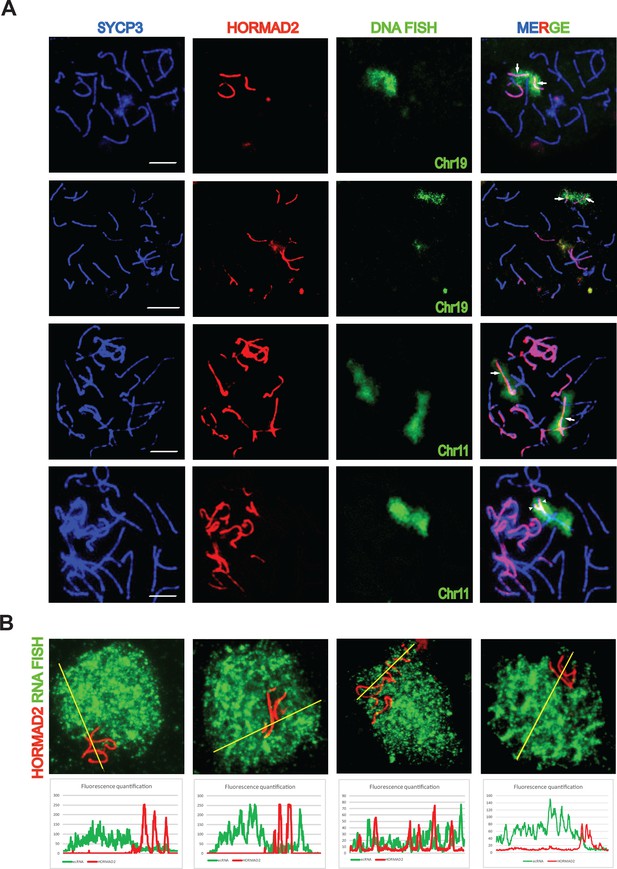
Asynapsis of heterosubspecific homologs in PB6F1 pachynemas.
(A) Partial (arrowheads) and complete (arrows) asynapsis of Chr 19 and 11. HORMAD2-labeled chromosomes with synapsis defects often form tangles via nonhomologous pairing. Scale bars represent 5 μm. (B) Asynapsed chromosomes are embedded in transcriptionally silenced chromatin visualized by the lack of extra-coding RNA (ecRNA) detected by Cot1 RNA FISH. See also Figure 1—video 1.
The images of the immunofluorescence anti-HORMAD2 stained and cot-1 RNA FISH-labeled spread spermatocytes were examined, and z-stack series were acquired using a confocal microscope (DMI6000CEL – Leica TCS SP8).
The acquired z-stack spectral series for HORMAD2 (red) and cot-1 RNA (green) were processed using the ImageJ program (https://imagej.net) to generate a 3D projection and a 360° video projection saved in the avi format.
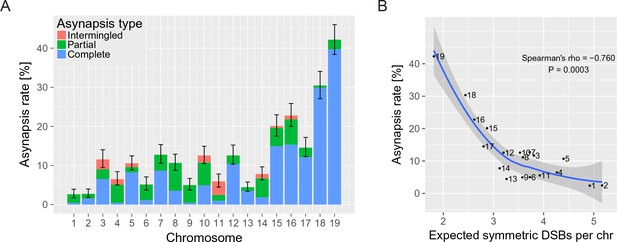
The asynapsis rate of individual autosomes in sterile male (PWD x B6)F1 hybrids.
(A) Mean asynapsis rate ±S.E (based on GLMM model). Intermingled asynapsis refers to overlaps of two or more asynapsed chromosomes within the DNA FISH cloud of chromatin. The five smallest chromosomes had higher asynapsis rate (GLMM model, p=1.1×10−13). Concurrently, the chromosomes with higher asynapsis rate were also more involved in complete rather than partial asynapsis (GLMM model, p=6.2×10−5). Proportion of complete and partial asynapsis was controlled by the asynapsis rate rather than by the chromosomal length (test for effect of the length when controlled for the asynapsis rate, p=0.491). (B) Negative correlation (Spearman’s ρ=−0.760, p=0.0003) between asynapsis rate and mean expected number of symmetric DSBs (Davies et al., 2016) based on the chromosome-wide distribution of SPO11 oligos in fertile B6 males (Lange et al., 2016).
-
Figure 2—source data 1
Asynapsis rate of individual chromosomes of (PWD x B6)F1 males.
- https://doi.org/10.7554/eLife.34282.006
-
Figure 2—source data 2
Chromosome-scale comparison of expected DSBs in symmetric hotspots and asynapsis rate.
- https://doi.org/10.7554/eLife.34282.007
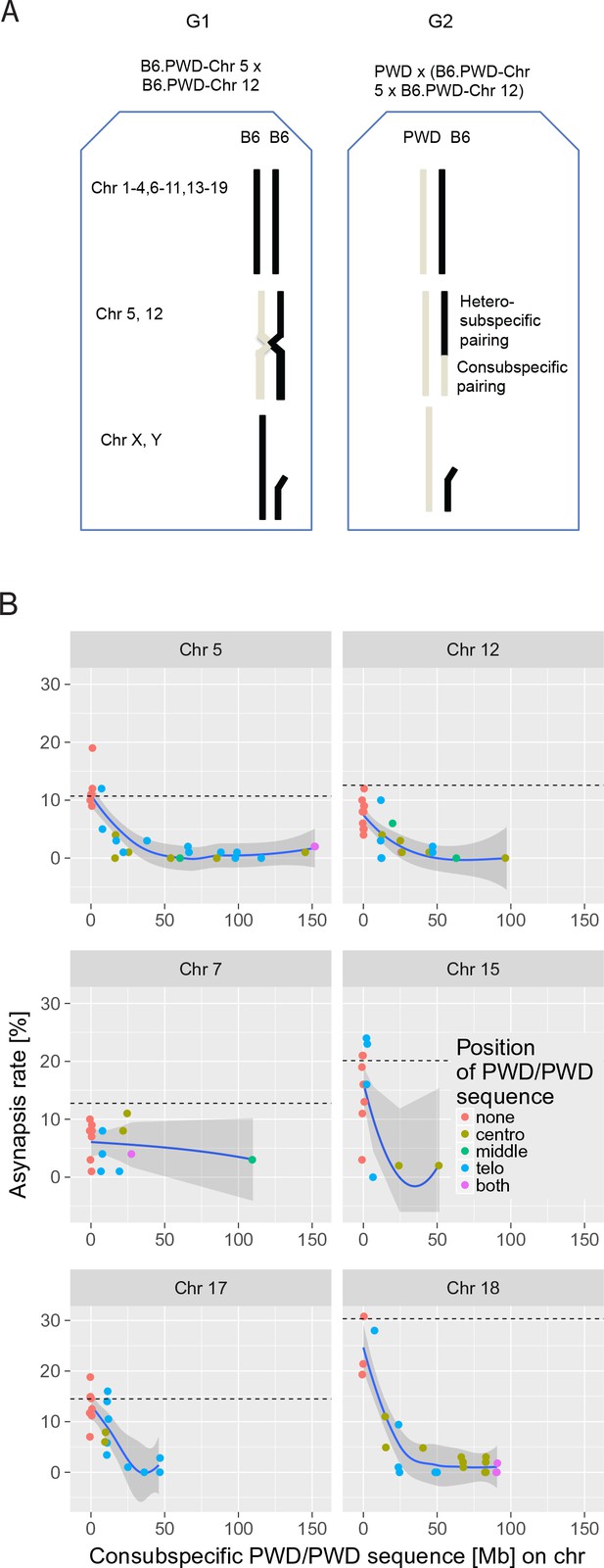
The effect of consubspecific PWD/PWD stretches of genomic sequence on pachytene synapsis, 2-chr cross.
(A) The F1 hybrid males of two consomic strains (generation 1 G1, Chr 5 and Chr 12 shown here) were crossed to PWD females to produce generation 2 G2 sterile F1 hybrids with random recombinant consomic chromosomes 5 and 12. Using whole-chromosome probes, the asynapsis rate of the consomic chromosomes was scored by DNA FISH. (B) Combination of two chromosomes (5 + 12, 7 + 15 and 17 + 18) were challenged in each experiment. The localization of PWD homozygous sequence with respect to centromere, interstitial part of the chromosome or telomere, or on both ends is distinguished by color (see also Figure 3—source data 1–6). The average length between the minimum and maximum of the consubspecific sequence is plotted. The mean asynapsis rate of a given chromosome is regularly higher in PB6F1 hybrids (dashed line) than in 2-chr cross. For explanation see Figure 4 and the section on the trans-effect-dependent variation in asynapsis rate. Loess curve with 95% CI.
-
Figure 3—source data 1
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 5.
- https://doi.org/10.7554/eLife.34282.009
-
Figure 3—source data 2
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 12.
- https://doi.org/10.7554/eLife.34282.010
-
Figure 3—source data 3
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 7.
- https://doi.org/10.7554/eLife.34282.011
-
Figure 3—source data 4
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 15.
- https://doi.org/10.7554/eLife.34282.012
-
Figure 3—source data 5
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 17.
- https://doi.org/10.7554/eLife.34282.013
-
Figure 3—source data 6
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chr 18.
- https://doi.org/10.7554/eLife.34282.014
-
Figure 3—source data 7
Change point estimates of the minimal length [Mb] of PWD/PWD homozygosity showing detectable affect on synapsis rate.
- https://doi.org/10.7554/eLife.34282.015
-
Figure 3—source data 8
Selected SSLP markers polymorphic between B6 and PWD.
- https://doi.org/10.7554/eLife.34282.016
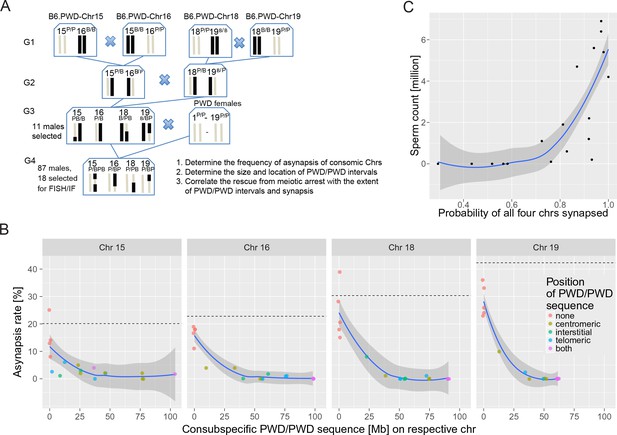
The effect of consubspecific PWD/PWD stretches of genomic sequence on pachytene synapsis and meiotic progression, 4-chr cross.
(A) Scheme of a four-generation cross resulting in F1 hybrids with four recombinant consomic chromosomes. (B) The asynapsis rate related to the size and chromosomal position of the consubspecific PWD/PWD sequence in four consomic chromosomes (15, 16, 18 and 19, see also Figure 4—source data 3). The localization of PWD homozygous sequence with respect to the centromere, the interstitial part of the chromosome, or the telomere, or on both ends is distinguished by color (see also Figure 4—source data 3). (C) Number of sperm in epididymis is a function of the probability of synapsis of all four consomic chromosomes. The complete meiotic arrest is reversed in males having 70% or higher chance of all four chromosomes synapsed. See Figure 6—figure supplement 2. Loess curve with 95% CI.
-
Figure 4—source data 1
Eleven G3 male parents selected for the 4-Chr cross experiment.
- https://doi.org/10.7554/eLife.34282.019
-
Figure 4—source data 2
The fertility parameters of hybrids of the 4-chr cross experiment.
- https://doi.org/10.7554/eLife.34282.020
-
Figure 4—source data 3
Four-chr cross.
The effect of the size and location of PWD/PWD consubspecific intervals on asynapsis of Chrs 15, 16, 18 and 19 and on fertility parameters.
- https://doi.org/10.7554/eLife.34282.021
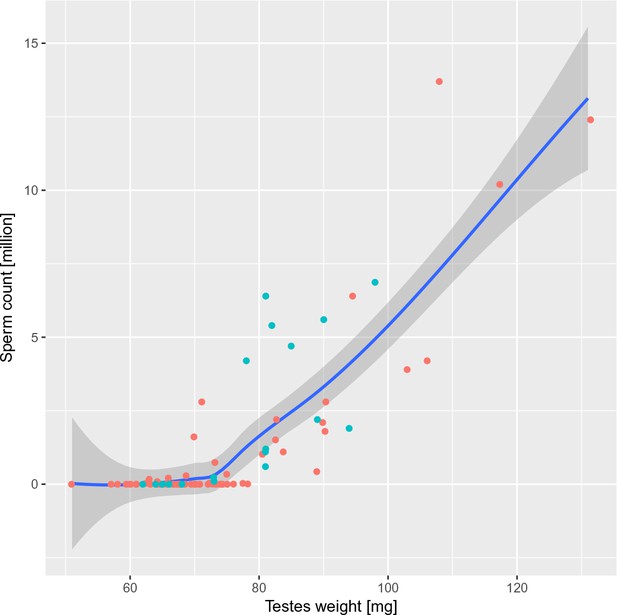
Fertility parameters of G4 males from the 4-chr cross.
Rescue of HS meiotic arrest is detectable in males with the weight of paired testes > 70 mg and >0.1×106 of sperm in the epididymis. The males selected for DNA FISH/HORMAD2 analysis of asynapsis are highlighted in turquoise. All males share the sterility-determining allelic combination of Prdm9PWD/B6 and Hstx2PWD. Loess curve with 95% CI.
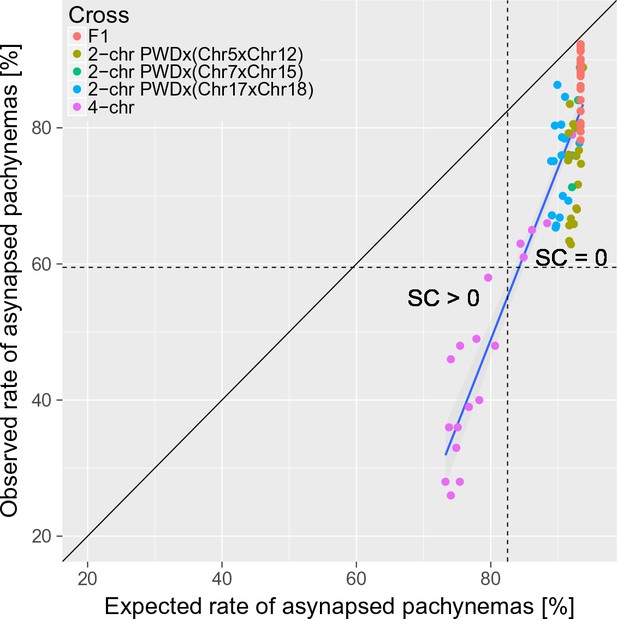
The trans-acting effect of consubspecific PWD/PWD stretches increases the probability of full synapsis of PWD/B6 intersubspecific homologs in males of 2-chr cross and 4-chr cross experiments.
The expected rate of synapsed pachynemas was calculated for each mouse in 2-chr cross and 4-chr cross experiments by multiplication of observed synapsis rates (i.e. assuming independence) of FISH analyzed chromosomes (e.g. Chrs 15, 16, 18 and 19 in 4-chr cross) and the observed PB6F1 synapsis rates of the remaining autosomes. Asynapsis rate was calculated as a complement to synapsis rate. The difference between expected and observed overall asynapsis is most pronounced in 4-chr cross males with the lowest expected overall asynapsis rate. Recovery of spermatogenesis signaled by the presence of sperm in the epididymis occurs when more than 40% of pachynemas are fully synapsed. SC is sperm count.
-
Figure 5—source data 1
Four-chr cross experiment.
The relation between observed and calculated rate of pachytene asynapsis and fertility parameters.
- https://doi.org/10.7554/eLife.34282.024
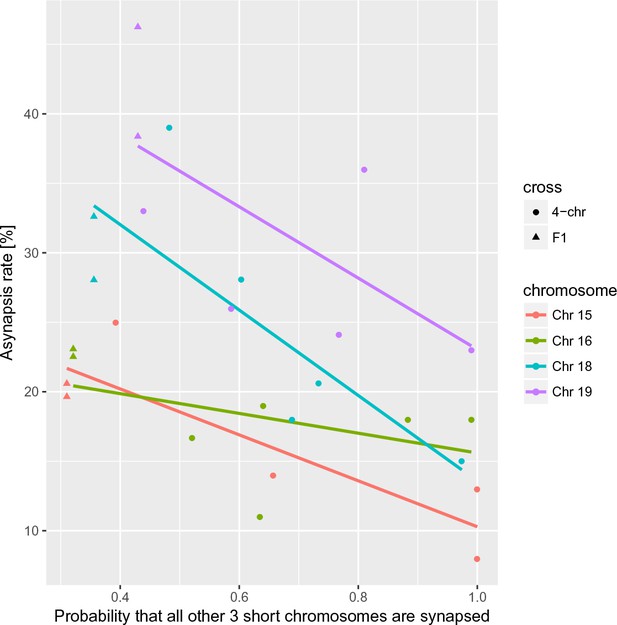
Asynapsis rate of individual nonrecombinant consomic PWD/B6 chromosomes modified in trans by the probability of synapsis of the remaining three consomic chromosomes in the 4-chr cross in individual males compared to PB6F1 hybrids.
Asynapsis rate of a given chromosome is in negative correlation (r = −0.88,–0.45, –0.80,–0.67 for Chrs 15, 16, 18, 19, respectively) with the probability that all of the other three analyzed chromosomes are synapsed. The estimate of the slope in log-log regression is β3_chrs_syn=–0.418 (95% CI = –0.272 to –0.534), overall p=0.0266.
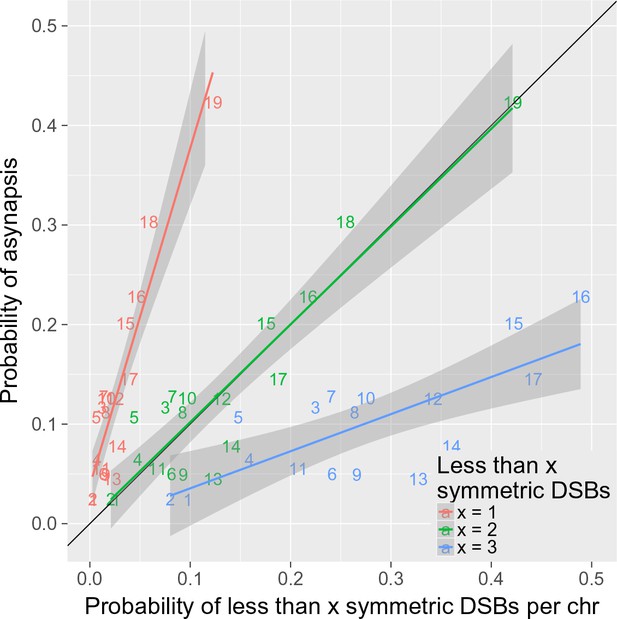
Two or more symmetric DSBs can be sufficient for synapsis.
The probability of less than one symmetric DSB per chromosome is ~4 times lower than the asynapsis rate observed in PB6F1 hybrid males (i.e. the estimate of probability of asynapsis), implying that ~75% of all asynapsis occurs when there is one or more repairable DSBs. The probability of less than two symmetric DSBs is a good estimate of the probability of asynapsis, whereas the probability of less than three symmetric DSBs overestimates the probability of asynapsis. This shows that, in the simplest explanation, two or more symmetric DSBs could be sufficient for synapsis. The probabilistic distribution of symmetric DSBs is calculated on the basis of the model described in the 'Discussion'.
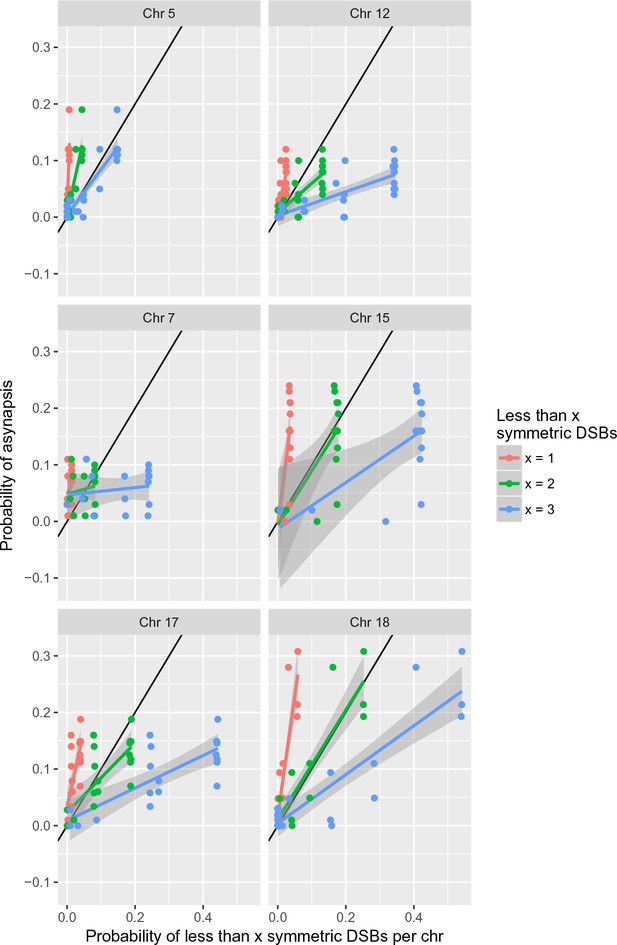
Two and more DSBs within symmetric hotspots can be sufficient for synapsis in 2-chr cross males.
See Figure 6 for details.
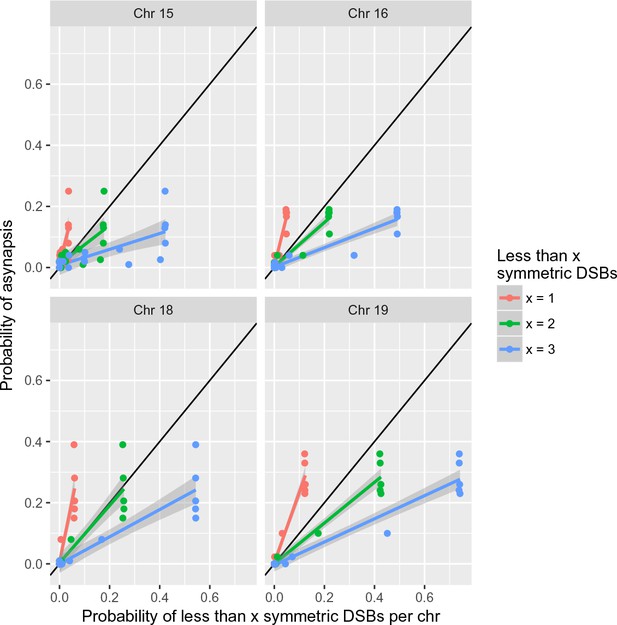
Two or more DSBs within symmetric hotspots can be sufficient for synapsis in 4-chr cross hybrid males.
See Figure 6 for details.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody - primary | Anti SYCP3 (mouse monoclonal, clone D-1) | Santa Cruz Biotechnology | sc-74569; SCP-3 Antibody (D-1); RRID:AB_2197353 | (1:50) |
| Antibody - primary | Anti gH2AFX (rabbit polyclonal) | Abcam | ab2893; gH2AFX antibody; RRID:AB_303388 | (1:1000) |
| Antibody - primary | Anti HORMAD2 (rabbit polyclonal) | DOI: 10.1371/journal.pgen.1000702 | Gift from Dr. Attila Toth | (1:700) |
| Antibody - primary | Anti HORMAD2 (rabbit polyclonal) | Santa Cruz Biotechnology | sc-82192; HORMAD2 antibody (C-18); RRID:AB_2121124 | (1:500) |
| Antibody - secondary | Anti-Rabbit IgG - AlexaFluor568 (goat polyclonal) | Molecular Probes | A-11036; RRID:AB_10563566 | (1:500) |
| Antibody - secondary | Anti-Mouse IgG - AlexaFluor647 (goat polyclonal) | Molecular Probes | A-21235; RRID:AB_2535804 | (1:500) |
| Blocking reagent forimmunostaining | Normal goat serum from healthy animals | Chemicon | S26-100ML | |
| Protease inhibitors | Complete, Mini, EDTA-free Protease Inhibitor Cocktail | Roche | 4693159001 | |
| Paraformaldehyde | Paraformaldehyde AQ solution | Electron Microscopy Sciences | 15714S | |
| DNA-FISH probes for mouse chromosomes1–19 | XMP X Green - Mouse chromosome paints | MetaSystems | D-1401–050-FI; D-1420–050-FI | |
| RNA FISH | Mouse Cot-1 DNA | Invitrogen | 18440016 | |
| RNA FISH | Biotin nick translation kit | Roche | 11,745,824,910 | |
| RNA FISH | Biotinylated goat anti-avidin antibody | Vector Laboratories | BA-0300; RRID:AB_2336108 | (1:100) |
| RNA FISH | Fluorescein-Avidin-DCS | Vector Laboratories | A-2011; RRID:AB_2336456 | (1:100) |
| RNase inhibitor | Ribonucleoside-vanadyl complex (RVC) | SIGMA - ALDRICH | 94742 | (1:100 = 2 mM) |
Additional files
-
Source code 1
The code in R contains simulations of assumed probabilistic distributions of numbers of symmetric DSBs per chromosome in F1, 2-chr and 4-chr crosses.
The probabilities of occurrence of respective numbers of symmetric DSBs are compared to the observed asynapsis rates in animals from F1, 2-chr and 4-chr crosses. This code enables reproduction of Figure 6, Figure 6—figure supplements 1 and 2.
- https://doi.org/10.7554/eLife.34282.028
-
Source code 2
This file, crosses_F1_2chr_4chr.csv, contains the data for Source Code File 1.
- https://doi.org/10.7554/eLife.34282.029
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34282.030





