Multi-view light-sheet imaging and tracking with the MaMuT software reveals the cell lineage of a direct developing arthropod limb
Figures
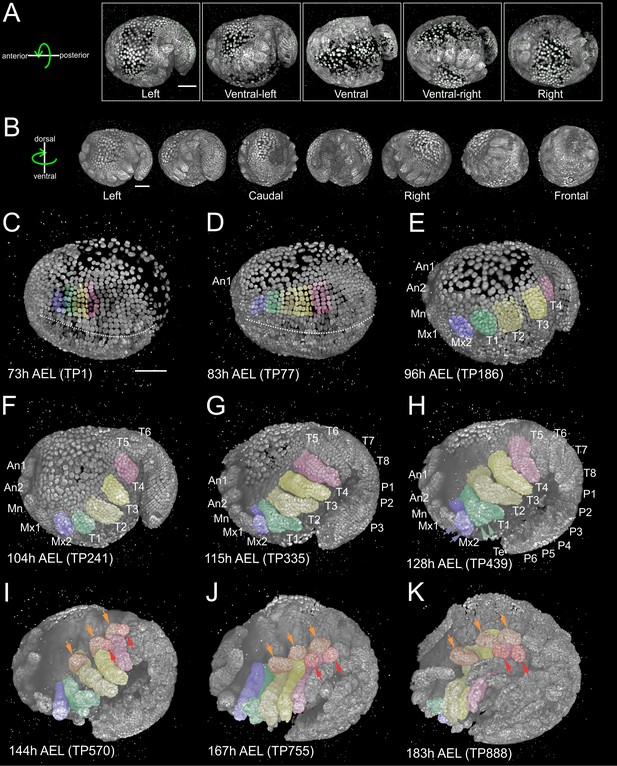
Reconstruction of Parhyale embryogenesis with multi-view LSFM (see also Figure 1—video 1 and 2).
(A) Transgenic Parhyale embryo with H2B-mRFPruby-labeled nuclei mounted with fluorescent beads (green dots) for multi-view reconstruction. The embryo was imaged from the indicated 5 views with 45˚ rotation around the AP axis between neighboring views. Panels show renderings of the acquired views with anterior to the left. (B) Raw views were registered and fused into an output image rendered here in different positions around the DV axis. (C–K) Each panel shows a rendering of the embryo at the indicated developmental stage in hours (h) after egg-lay (AEL) and the corresponding time-point (TP) of the recording. Anterior is to the left and dorsal to the top. Abbreviations: first antenna (An1), second antenna (An2), mandible (Mn), maxilla 1 (Mx1), maxilla 2 (Mx2), thoracic appendages 1 to 8 (T1–T8), pleonic (abdominal) appendages 1 to 6 (P1–P6) and telson (Te). Color masks indicate the cells contributing to Mx2 (blue), T1 (green), T2 and T3 (light and dark yellow) and T4 limb (magenta). (C) Embryo at mid-germband stage S13 according to (Browne et al., 2005). The ventral midline is denoted with the dotted line. (D) S15 embryo. Germband has extended to the posterior egg pole and the first antennal bud is visible anteriorly. (E) S18 embryo with posterior flexure. Head and thoracic appendages have bulged out up to T4. (F) S19 embryo with prominent head and thoracic appendage buds up to T6. (G) S20 embryo continues axial elongation ventrally and anteriorly. Appendage buds are visible up to P3. (H) S21 embryo. Segmentation is complete and all appendages have formed. The Mx2 has split into two branches (blue arrowheads) and the T1 limb has developed two proximal ventral outgrowths (green arrowheads). (I) Embryo at stage S22, (J) S23, and (K) S24 showing different phases of appendage segmentation. Dorsal outgrowths at the base of thoracic appendages, namely coxal plates (orange arrowheads) and gills (red arrowheads), are indicated in T2, T3 and T4. Scale bars are 100 µm.
Imaging Parhyale embryogenesis with multi-view LSFM
Time-lapse recording of a transgenic embryo from the crustacean amphipod Parhyale hawaiensis labeled with the nuclear H2B-mRFPruby fluorescent marker. The embryo was recorded on a Zeiss Lightsheet Z.1 microscope that offers two-sided illumination and single-sided detection with a 20x/1.0 objective. This embryo was imaged from 5 views 45˚ apart (ventral side, the two ventral-lateral sides and two lateral sides) for slightly longer than 4.5 days with a temporal resolution of 7.5 min. Development of the entire embryo was reconstructed from the five input views by registering them in space and time using fluorescent beads scattered in the agarose where the embryo was embedded, by fusing the registered views into a single output image with a more isotropic resolution using a multi-view deconvolution algorithm, and by rendering of the fused volume and rotating it around the anterior-posterior x-axis over time. All image-processing steps were carried out with open-source software available in the Fiji image analysis platform. The movie plays 3 hr of Parhyale development per second, displaying development 10800 times faster than normal. The movie starts at 3 days AEL, when a distinct germband has formed ventrally and is surrounded by large spaced-out nuclei of extra-embryonic identity. Segment formation and maturation progresses from anterior to posterior and is accompanied by embryo elongation, first posteriorly and later on ventrally and anteriorly. During these stages, the embryo develops a series of specialized appendages along the anterior-posterior axis that can be observed projecting ventrally, elongating and segmenting along their proximal-distal axis. Anterior is to the left.
Imaging Parhyale embryogenesis with multi-view LSFM
Left side of the same embryo shown in Figure 1—video 1 rendered with the same settings but without rotation. Anterior is to the left and dorsal to the top.
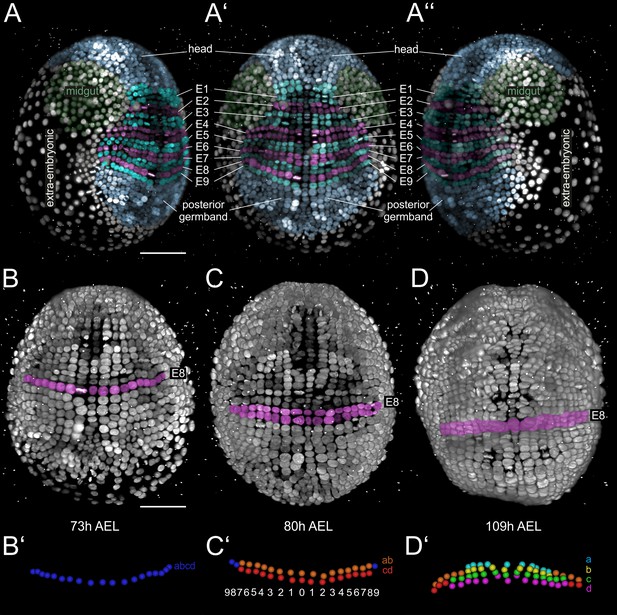
Grid architecture of the Parhyale germband.
(A–A’’) Rendering of a Parhyale embryo at the growing germband stage: (A) Right, (A’) ventral, and (A’’) left side. Color masks indicate the anterior head region (blue), the bilaterally symmetric midgut precursors (green), the orderly arranged parasegments E1 to E9 (in alternating cyan and magenta), the posterior end of the germband with ongoing organization of cells into new rows (blue), and the extra-embryonic tissue (white). (B–D) Ventral views of elongating germband at the indicated hours (h) after egg-lay (AEL). Ectodermal cells of the E8 parasegment are shown in magenta. (B’) Schematics of tracked E8 abcd cells (blue) in the 1-row-parasegment, (C’) anterior ab cells (orange) and posterior cd cells (red) after the first longitudinally-oriented division in the 2-row-parasegment, and (D’) a (cyan), b (yellow), c (green) and d cells (magenta) after the second longitudinally-oriented division in the 4-row-parasegment. Both mitotic waves proceed in medial-to-lateral direction. The resulting daughter cells sort in clearly defined columns that are identified by ascending index numbers with 0 denoting the ventral midline and 1, 2, 3 etc. the more lateral columns with increasing distance from midline.
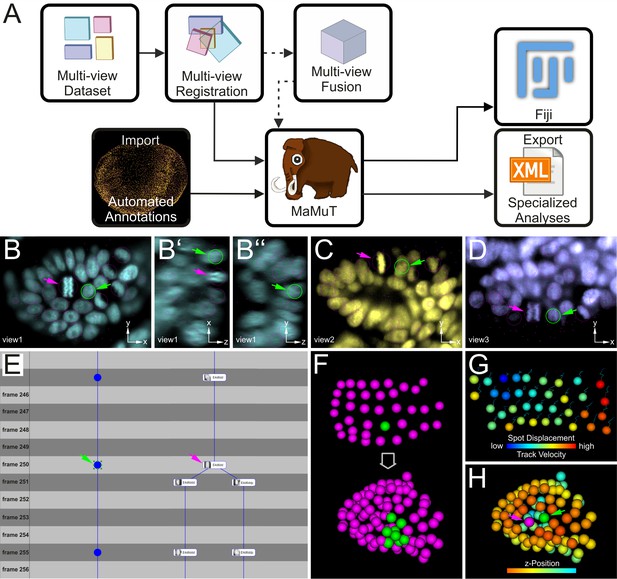
Cell tracking and lineage reconstruction with MaMuT (see also Figure 3—figure supplement 1).
(A) Workflow for image data analysis with MaMuT. Raw views (colored boxes in Multi-view Dataset) are registered (overlapping boxes in Multi-view Registration) and, optionally, fused into a single volume (large cube in Multi-view Fusion). The raw (and fused) image data together with the registration parameters are imported into MaMuT (mammoth logo). In its simplest implementation, all data analysis is done with MaMuT in Fiji workspace. In more advanced implementations, automated segmentation and tracking annotations (yellow point cloud of tracked cells) can be computed separately and imported into MaMuT. The reconstructed lineage information can be exported from MaMuT in an xml file for specialized analyses in other platforms. (B–D) The MaMuT Viewer windows display the raw image data and annotations. All tracked nuclei are marked with magenta circles (in view) or dots (out of view). The active selection is marked in green in all synced Viewers: (B) xy, (B’) xz, and (B’’) yz plane of first view in cyan; (C) xy plane of second view in yellow; (D) xy plane of third view in blue. (E) The TrackScheme lineage browser and editor where tracks are arranged horizontally and time-points vertically. Tracked objects can be displayed simply as spots (left track) or with extra information like their names and thumbnails (right track). Tracks are displayed as vertical links. The TrackScheme is synced with the Viewer windows; the selected nucleus in panels B–D is also highlighted here in green at the indicated time-point (called frame). Objects can be tracked between consecutive time-points or in larger steps. (F–H) The 3D Viewer window displays interactive animations of tracked objects depicted as spheres. Spots and tracks can be color-coded by lineage, position and other numerical parameters extracted from the data. (F) Digital clone of a nucleus (shown in green) tracked from the grid stage to the limb bud stage. All other tracked nuclei are shown in magenta. (G) Spots color-coded by displacement and tracks color-coded by velocity. (H) Tracked nuclei in the limb bud mapped out in different colors based on z-position. In panels B–E and H, the selected nucleus and the neighboring dividing nucleus are indicated with green and magenta arrowheads, respectively.

MaMuT layout.
(A–C) The three tabs of the MaMuT control panel. (A) The Views tab is used to launch and control the different displays of the image data and annotations. (B) The Annotation tab is used to define the temporal sampling during manual lineaging and the parameters for semi-automated tracking. (C) The Actions tab allows users to generate movies of tracked objects or merge independent MaMuT annotations of the same image dataset into a single file. (D) The Visibility and Grouping panel allows users to organize views into groups and display them overlaid in the same Viewer window. (E) The Brightness and Color panel is used to adjust brightness, contrast and color of the image data in the Viewer windows. For example, three different views are shown in panels G-L in cyan, blue and yellow, respectively. (F) The MaMuT help menu with the default mouse and keyboard operations that can be modified by the user. (G–L) The MaMuT Viewer windows display the raw image data. The user interacts with the data to create and edit annotations through these Viewers and the TrackScheme window. The user can open as many Viewer windows as required for accurate tracking, and can display the image data in any useful scale, position and orientation (two orientations shown here per view). (M) The TrackScheme window is the dedicated lineage browser and editor where tracks are arranged from left to right and time-points from top to bottom. (N) The 3D Viewer window shows animations of the tracked objects as spheres without the image data. The TrackScheme, the 3D Viewer and all open Viewer windows are synced (with the active selection highlighted in bright green) and annotations can be color-coded according to various parameters extracted from the data.
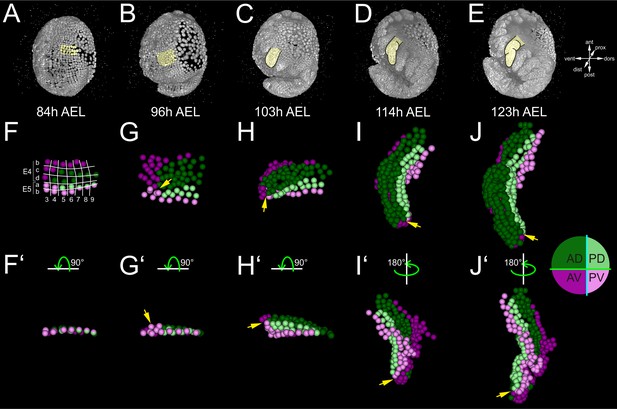
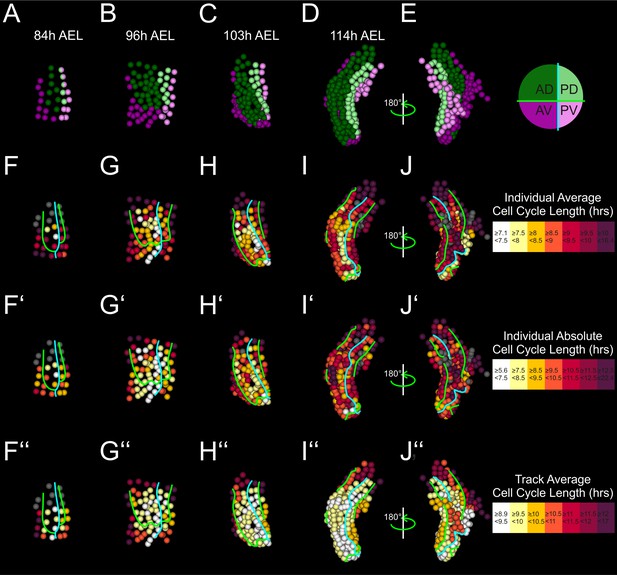
Early compartmentalization of the Parhyale thoracic limb (see also Figure 4—figure supplements 1 and 2).
(A–E) Lateral views of a Parhyale embryo rendered at the indicated developmental stages shown in hours (h) after egg-lay (AEL). Yellow masks show the left T2 limb (limb#1). (F–J’) Tracked cells contributing to limb#1 were color-coded by their compartmental identity: Anterior-Dorsal (dark green), Anterior-Ventral (dark magenta), Posterior-Dorsal (light green), and Posterior-Ventral (light magenta). (F) Ventral view of limb primordium at 84 hr AEL made up by cells from the E4 and E5 parasegments. Horizontal lines separate AP rows a to d and vertical lines separate DV columns 3 to 9. (F’) Posterior view, rotated 90˚ relative to F. (G) Ventral view of the limb during early eversion at 96 hr AEL. (G’) Posterior view, rotated 90˚ relative to G. The cells close to the intersection of the four compartments (yellow arrows) are the first to rise above the level of the epithelium. (H–J) Dorsal views of (H) limb bud at 103 hr AEL, (I) initial limb elongation at 114 hr AEL and (J) later elongation phase at 123 hr AEL. (H’) Posterior view, rotated 90˚ relative to H, and (I’–J’) ventral views, rotated 180˚ relative to I-J. The intersection of the AP and DV boundaries (yellow arrows) is located at the tip of the limb.
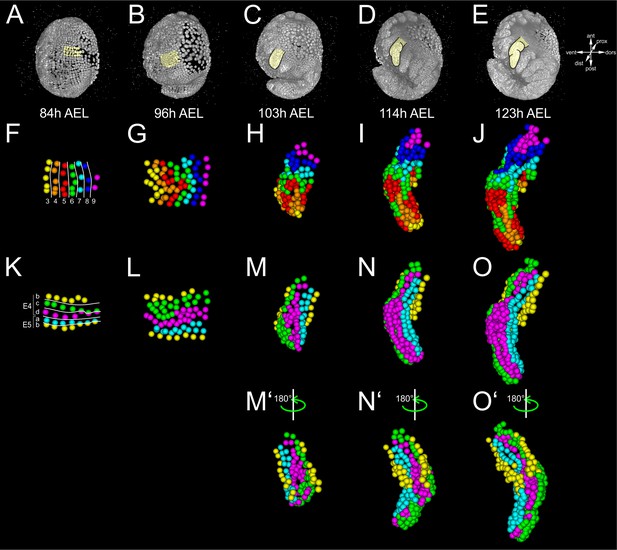
Lineage reconstruction of the Parhyale thoracic limb.
(A–E) Lateral views of the same Parhyale embryo shown in Figure 4. Yellow masks indicate the left T2 limb (limb#1). (F–J) Tracked cells contributing to the T2 limb color-coded by the DV column they belong to: column three in yellow, four in orange, five in red, six in green, seven in cyan, eight in blue, and nine in magenta. Note the cell mixing and irregular clone borders between descendent cells from neighboring columns. (K–O’) Tracked cells contributing to the T2 limb belong to the E4 and E5 parasegments, and are color-coded by the AP row they belong to: a row in cyan, b rows in yellow, c row in green and d row in magenta. Note the absence of cell mixing at the AP compartment boundary between anterior E4d cells (magenta) and posterior E5a cells (cyan), as well as the absence of cell mixing at the presumptive DV boundary between ventral E4b cells (yellow) and dorsal E4c cells (green) anteriorly, and between ventral E5b cells (yellow) and dorsal E5a cells (cyan) posteriorly. (F,K) Ventral views of the limb primordium at 84 hr (h) after egg-lay (AEL). (G,L) Ventral views of the limb during early eversion at 96 hr AEL. (H,M) Dorsal views of the limb bud at 103 hr AEL. (I,N) Dorsal views of initial limb elongation at 114 hr AEL. (J,O) Dorsal views of later elongation phase at 123 hr AEL. (M’–O’) Ventral views, rotated 180˚ relative to M–O.

Independent evidence for early compartmentalization of the Parhyale thoracic limb.
(A–D) Lateral views of another Parhyale embryo imaged on a Zeiss LSFM prototype instrument rendered at the indicated developmental stages shown in hours (h) after egg-lay (AEL). Yellow masks indicate the left T2 limb (limb#2). (E–H’) Cells contributing to limb#2 were tracked with the SIMI°BioCell software and were color-coded by their compartmental identity: Anterior-Dorsal (dark green), Anterior-Ventral (dark magenta), Posterior-Dorsal (light green), and Posterior-Ventral (light magenta). (E) Ventral view of the limb primordium at 86 hr AEL. Horizontal lines separate AP rows a to d, and vertical lines separate DV columns 3 to 9. (E’) Posterior view, rotated 90˚ relative to E. (F) Ventral view of the limb primordium during eversion at 94 hr AEL. (F’) Posterior view, rotated 90˚ relative to F. The cells close to the intersection of the four compartments (yellow arrows) have risen above the level of the epithelium. (G–H) Dorsal views of limb bud at 98 hr AEL and initial limb elongation at 101 hr AEL. (G’–H’) Ventral views, rotated 180˚ relative to G–H. The intersection of the AP and DV compartment boundaries (yellow arrows) marks the tip of the limb. Limb#2 exhibited the same lineage restrictions like the more completely reconstructed limb#1. Cells in the anterior compartment (E4b, c and d rows) remained together and separate by a straight boundary from the cells in the posterior compartment (E5a and b rows). Likewise, no cell mixing was detected across the dorsal-ventral compartment boundary that extended again between the E4b and c rows anteriorly, between the E5a and b rows posteriorly, and between cells E4c4-c5, E4d3-d4 and E5a4-a5 medially.
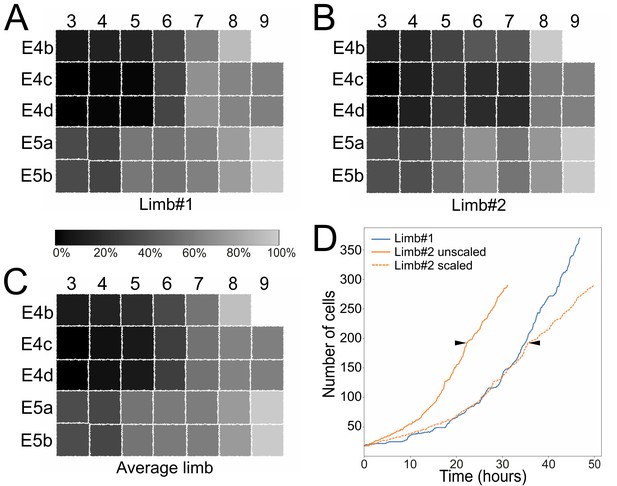
Stereotyped and variable cell behaviors in developing Parhyale thoracic limbs (see also Figure 5—figure supplement 1, Figure 5—source data 1, and Figure 5—video 1).
(A–C) Schematic representations of the T2 limb primordium at the 4-row-parasegment stage displaying the 34 founder cells as squares color-coded based on their relative birth times: (A) limb#1, (B) limb#2 and (C) their average. The first forming E4c3/d3 cells are colored in black (0% birth time difference), the last forming E5a9/b9 cells in light gray (100% birth time difference) and all other cells in intermediate grayscale shades based on their birth time difference relative to E4c3/d3. (D) Change in cell number over time in limb#1 (blue line) imaged at 26˚C and in the faster developing limb#2 (orange lines) imaged at 29–30˚C. The first division of the E4cd3 cell is the starting point for both growth curves. Solid lines show the raw data for the two limbs, while the dashed orange line shows the temporally registered data for limb#2. Arrowheads indicate the unscaled and scaled time-point up to which the SIMI°BioCell reconstruction of limb#2 was complete. An increasing number of cells in limb#2 were not possible to track after this time-point resulting in a poor registration with the growth curve of limb#1.
-
Figure 5—source data 1
Relative birth times of founder cells in Parhyale thoracic limbs.
- https://doi.org/10.7554/eLife.34410.013
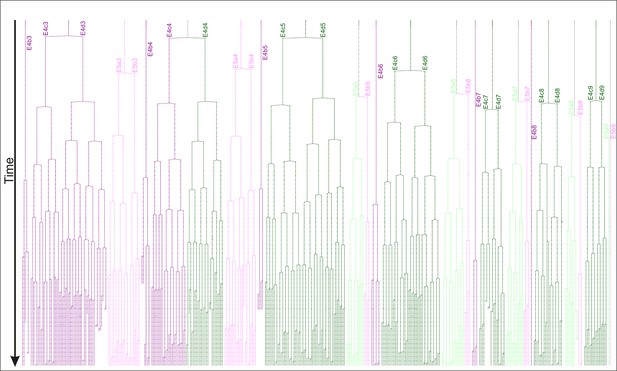
Reconstructed lineage tree of a Parhyale T2 limb.
Each track resembles one or two of the 34 founder cells of limb#1 color-coded by their compartmental identity: anterior-dorsal in dark green, anterior-ventral in dark magenta, posterior-dorsal in light green, and posterior-ventral in light magenta. Tracks labeled with the names of the 34 cells are arranged horizontally and the 400 time-points of tracking time, corresponding to 50 hr of development, are arranged vertically.
Animation of tracked cells forming the Parhyale second thoracic limb
All tracked cells contributing to the T2 limb#1 are displayed as spheres of uniform color. The movie starts from the early limb specification stage at about 3 days AEL and covers limb bud formation and initial elongation over the following 50 hr. This animation was produced with the 3D Viewer of the Massive Multi-view Tracker (MaMuT) plugin in Fiji. The movie plays 5 hr of limb development per second, displaying Parhyale limb development 18000 times faster than normal.
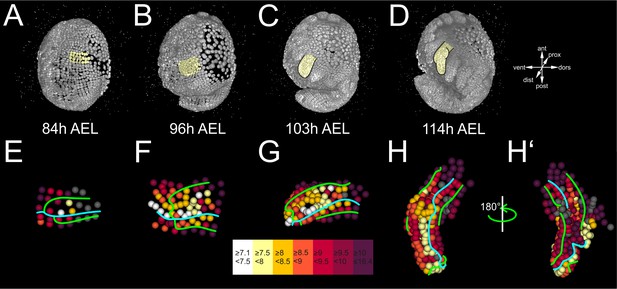
Differential cell proliferation rates in the Parhyale thoracic limb (see also Figure 6—figure supplements 1 and 2).
(A–D) Lateral views of the same Parhyale embryo shown in Figure 4. (E–H’) Tracked cells in limb#1 were color-coded by their average cell cycle length according to the scale (in hours) shown at the bottom. AP and DV boundaries are indicated by the cyan and green line, respectively. Cells for which measurements are not applicable are shown in gray. (E) Ventral view of the limb primordium at 84 hr (h) after egg-lay (AEL). Some central c and d cells start dividing faster at the 4-row-parasegment. (F) Ventral view of the limb during early eversion at 96 hr AEL with the middle cells dividing faster than peripheral cells. (G) Dorsal view of limb bud at 103 hr AEL. Higher proliferation rates are detected at the tip and in the anterior-dorsal compartment. (H) Dorsal and (H’) ventral view of elongating limb at 114 hr AEL. Cells at the tip of the limb and anterior cells abutting the AP compartment boundary divide the fastest.
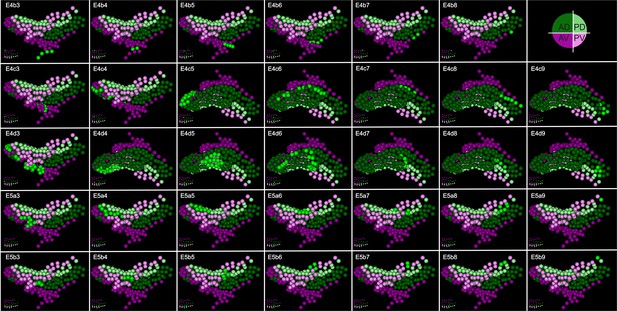
Digital clonal analysis in the Parhyale thoracic limb.
Digital clones for each one of the 34 founder cells of the T2 limb#1 visualized at 114 hr (h) after egg-lay (AEL). In each panel, the name of the founder cell is shown in the top left corner and its position in the limb primordium at 84 hr AEL is shown in the bottom left corner. The cells of the clone are shown in bright green. The rest cells have been color-coded by their compartmental identity: Anterior-Dorsal in dark green, Anterior-Ventral in dark magenta, Posterior-Dorsal in light green, and Posterior-Ventral in light magenta.
Alternative quantifications of cell proliferation rates in the Parhyale thoracic limb.
(A –E) Tracked cells making up the T2 limb#1 shown at the indicated hours (h) after egg-lay (AEL) and color-coded by their compartmental identity: Anterior-Dorsal in dark green, Anterior-Ventral in dark magenta, Posterior-Dorsal in light green, and Posterior-Ventral in light magenta. (F–J) Same stages as in A–E with cells color-coded by the average cell cycle length of each cell according to the scale shown on the right. (F’–J’) Same stages as in A–E with cells color-coded by the absolute cell cycle length of each cell according to the scale shown on the right. (F’’–J’’) Same stages as in A–E with cells color-coded by the average cell cycle length of each track according to the scale shown on the right. The AP and DV compartment boundaries are indicated by the cyan and green line, respectively. During the early stages, all analyses - irrespective of the method of quantification - demonstrate that a group of central cells in the limb primordium divide faster compared to peripheral cells. During later stages, the higher cell proliferation rates at the tip of the limb and at the AP compartment boundary are more pronounced with the calculation of the average cell cycle length for each cell. Gray cells indicate cells for which measurements are not applicable.
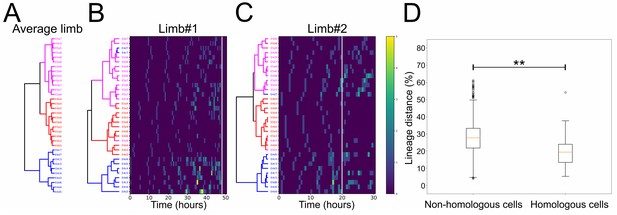
Lineage comparisons within and across Parhyale thoracic limbs (see also Figure 7—source data 1).
(A) Hierarchical clustering of the 34 founder cells in the Parhyale T2 limb based on a distance matrix computed from their average division patterns in limb#1 and limb#2. The cluster of E4c3-c7 and E4d3-d7 cells at the bottom is shown in blue, the middle cluster containing primarily the E4 and E5 b cells is shown in red, and the top cluster with the remaining cells is shown in magenta. (B,C) Hierarchical clustering of the 34 founder cells in (B) limb#1 and (C) limb#2 based on distance matrices computed from the division patterns observed in each limb. The cells in the two trees (color-coded as in A) display very similar clustering profiles. Heat maps show the timing and number of divisions in five time-point-windows. For each founder cell, divisions are represented with rectangles color-coded according to the number of divisions shown in the color bar. The x-axis shows the unscaled tracking time for each limb starting from division of the E4cd3 cell and the white line indicates the time-point at which cells were compared. (D) Box plots showing the distribution of lineage distances in pairwise comparisons between non-homologous (left) and homologous (right) founder cells across the two limbs. The two distributions differ significantly at p≤0.01 based on the Kolmogorov-Smirnov test. The data used for limbs #1 and #2 in panels A and D were at a comparable stage of their development indicated with the arrowheads in Figure 5D.
-
Figure 7—source data 1
Lineage distances between founder cells in Parhyale thoracic limbs.
- https://doi.org/10.7554/eLife.34410.020
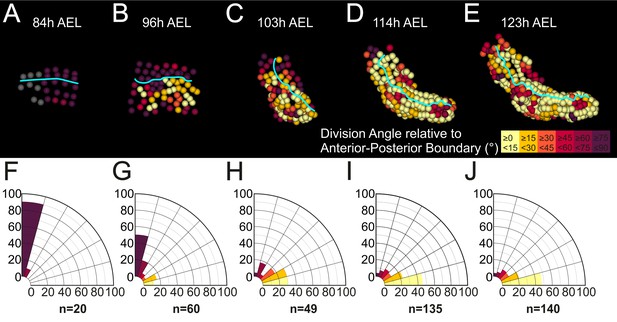
Oriented cell divisions in the Parhyale thoracic limb.
(A–E) Cells in the T2 limb#1 shown at the indicated hours (h) after egg-lay (AEL) color-coded by the orientation of mitotic divisions relative to the AP boundary (cyan line). The AP boundary is parallel to and an accurate proxy for the PD axis during limb outgrowth. The absolute values of the division angle relative to the AP boundary are sorted in 6 bins of 15˚. Gray cells in panel A indicate non-divided cells. (F–J) Rose diagrams with 15˚ intervals showing the percentage of mitotic events falling in each bin color-coded as in A-E (n shows the actual number of divisions). (A,F) Only longitudinally-oriented divisions (perpendicular to the AP boundary) are detected in the limb primordium 73 to 84 hr AEL. (B,G) Most cells still divide longitudinally 84 to 96 hr AEL, but an increasing number of dividing cells align parallel to the AP boundary during early eversion. (C,H) More than 59% of cells divide 0˚−30˚ relative to the AP boundary in the limb bud from 96 to 103 hr AEL. (D,I) Early and (E,J) later limb elongation phase from 103 to 123 hr AEL with the large majority of cells (>68%) dividing 0˚−30˚ relative to the AP boundary.
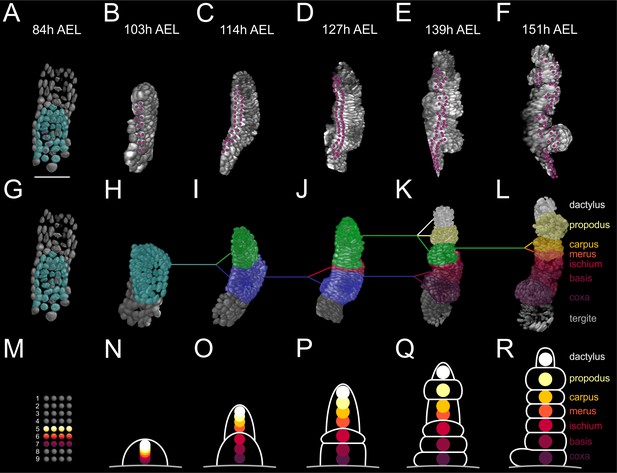
Elaboration of the Parhyale limb PD axis (see also Figure 9—figure supplement 1).
(A–F) Rendering of the T2 limb#1 at the indicated hours (h) after egg-lay (AEL). The cells contributing to the T2 primordium are shown in cyan in panel A. Magenta dots indicate the tracked cells E5a5-a8 and their descendants. Panel A shows a ventral view of the germband and panels B–F posterior views of the T2 limb. (G–L) Same stages as in A–F with color masks showing (G) the limb primordium, (H) the early limb bud, (I) the 2-partite limb with the first subdivision between ischium/merus, (J) the 3-partite limb after the second subdivision between basis/ischium, (K) the 6-partite limb after three more subdivisions between coxa/basis, propodus/dactylus and carpus/propodus, and (L) the final pattern made of 7 segments after the carpus/merus division. Colored lines indicate the relationships between limb parts in consecutive stages. (M–R) Schematics of limb subdivisions along the PD axis at the same time-points as in panels G–L. The rectangular lattice in panel M shows the 9 columns of cells in the 4-row-parasegment. White lines in panels N–R delineate the subdivisions of the T2 limb. The origin of each of the seven limb segments is shown with discs color-coded by segment.

Proximal-distal lineage separation in the growing Parhyale thoracic limb.
(A–E) Tracked cells contributing to the T2 limb#1 color-coded by their compartmental identity: Anterior-Dorsal (dark green), Anterior-Ventral (dark magenta), Posterior-Dorsal (light green), and Posterior-Ventral (light magenta). (A) Ventral view of the limb primordium at 84 hr (h) after egg-lay AEL. (B) Ventral view of the limb during early eversion at 96 hr AEL. (C) Dorsal view of limb bud at 103 hr AEL. Posterior views of (D) initial limb elongation at 114 hr AEL and (E) later elongation phase at 123 hr AEL. (F–J) Same stages and views as in A-E with cells contributing to the proximal (p) leg segments (coxa, basis, and ischium) shown in cyan and cells contributing to the distal (d) leg segments (merus, carpus, propodus, and dactylus) shown in yellow. Progenitor cells giving rise to both proximal and distal leg segments are shown in bright green. (K–M) Later stages of limb segmentation at (K) 132 hr AEL, (L) 140 hr AEL and (M) 150 hr AEL. In these panels, the T2 limb has been rendered in posterior view and superimposed with the tracked cells (descendant cells from posterior-dorsal progenitors E5a5-a8 shown as dots) covering the limb proximal-distal axis. Note that the proximal cells (in cyan) and the distal cells (in yellow) stop mixing at the ischium/merus joint (demarcated with the white line in K–M) after about 110 hr AEL. (N) Cell lineage tree of limb#1 where the tracks have been color-coded by their proximal or distal identity: proximal identity in cyan, distal identity in yellow, and mixed identity in green. Proximal and distal cells originate from the peripheral and medial territories of the limb primordium, respectively.
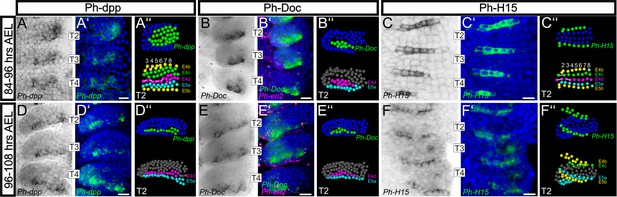
Analysis of developmental regulatory genes corroborates cellular models of limb morphogenesis (see also Figure 10—figure supplement 1).
(A–F) Brightfield images of T2, T3 and T4 limbs from S16-S18 embryos (top row, 84–96 hr AEL) and S19 embryos (bottom row, 96–108 hr AEL) stained by in situ hybridization for Ph-dpp (left columns), Ph-Doc (middle columns) and Ph-H15 (right columns). (A’–F’) Same limbs as in panels A–F with the nuclear DAPI staining in blue overlaid with the Ph-dpp, Ph-Doc or Ph-H15 pattern false-colored in green. Embryos stained for Ph-Doc were co-hybridized with Ph-en2 shown in magenta to label the posterior compartment. (A’’–F’’) MaMuT reconstructions of the T2 limbs shown in panels A–F. The top panels are color-coded by gene expression with Ph-dpp, Ph-Doc or Ph-H15 expressing cells shown in green and non-expressing cells in blue. Bottom panels indicate the identity of the same cells; cells are color-coded by AP rows, column number is shown at the top and white lines connect sister cells. All panels show ventral views with anterior to the top and ventral midline to the left. Scale bars are 20 μm.
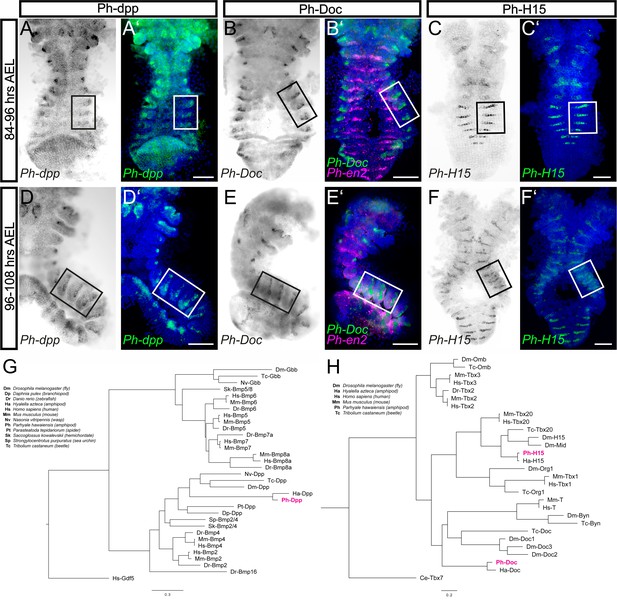
Expression of Ph-dpp, Ph-Doc and Ph-H15 during Parhyale limb bud formation.
(A–F) Brightfield images of S16-S18 embryos (top row, 84–96 hr AEL) and S19 embryos (middle row, 96–108 hr AEL) stained by in situ hybridization for Ph-dpp (left columns), Ph-Doc (middle columns) and Ph-H15 (right columns). (A’–F’) Same embryos as in panels A–F with the nuclear DAPI staining in blue overlaid with the Ph-dpp, Ph-Doc or Ph-H15 pattern false-colored in green. Embryos stained for Ph-Doc were co-hybridized with Ph-en2 shown in magenta to label the posterior compartment. Panels show ventral views with anterior to the top. Rectangles indicate the T2, T3 and T4 limbs shown in Figure 10. Scale bars are 100 μm. (G) Phylogenetic analysis of BMP family proteins and (H) T-box transcription factors. Scale bars show number of substitutions per site.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Parhyale hawaiensis) | Wild Type | PMID: 15986449 | ||
| Strain, strain background (P. hawaiensis) | PhHS>H2B-mRFPruby | This paper | ||
| Recombinant DNA reagent | pMi{3xP3>EGFP; PhHS>H2B-mRFPruby} | This paper | ||
| Software, algorithm | MaMuT | This paper | http://imagej.net/MaMuT | |
| Software, algorithm | SIMI°BioCell | PMID: 9133433 | http://simi.com/en/products/cell-research | |
| Gene (P. hawaiensis) | Ph-dpp | This paper | GenBank: KY696711 | |
| Gene (P. hawaiensis) | Ph-Doc | This paper | GenBank: KY696712 | |
| Gene (P. hawaiensis) | Ph-en2 | This paper | GenBank: KY696713 | |
| Gene (P. hawaiensis) | Ph-H15 | This paper |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.34410.026





