IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis
Figures
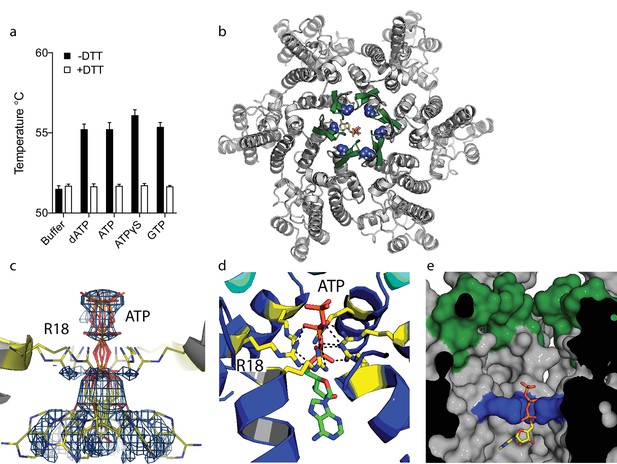
ATP binds to the HIV-1 capsid.
(a) Change in capsid hexamer stability upon addition of different nucleotide triphosphates as measured by differential scanning fluorimetry. Data are from three replicates and representative of at least three independent experiments. (b) Complexed structure of ATP bound to HIV capsid hexamer. The N-terminal β-hairpin is coloured in green, the location of R18 in blue spheres and ATP as bonds. (c) Electron density for ATP in the complex. (d) R18 in the HIV-1 capsid interacts with the adenosine triphosphates. (e) A cutaway view of the HIV-1 capsid surface in cross-section showing ATP (as sticks) bound inside the hexamer. The β-hairpin is in green and R18 in blue.
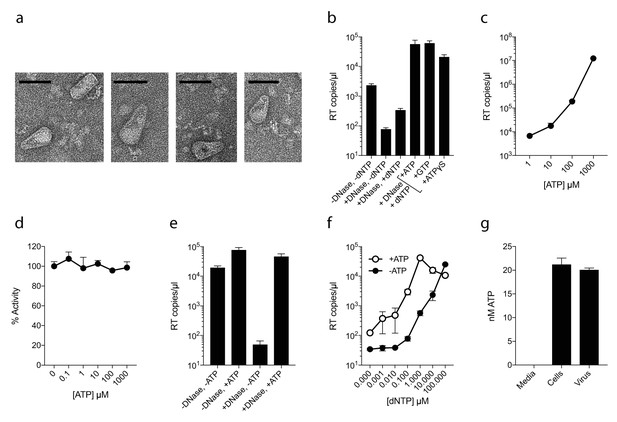
ATP does not prevent nucleotide import and ERT.
(a) Electron micrographs of isolated HIV-1 cores as visualised by negative staining. Size bars are 100 nm. (b) ERT in presence of nucleotide triphosphates. (c) ATP dose-dependently increases ERT efficiency. (d) ATP has no effect on in vitro RT in the absence of a capsid. (e) ATP increases accumulation of encapsidated RT products in the presence of DNase. (f) ATP increases ERT at dNTP concentrations < 100 µM. (g) ATP levels in producer cells (~100 cells) and HIV-1 virions (~5×109 virions) as detected by luciferase assay (see Materials and methods). All data are mean of three replicates ± SD.
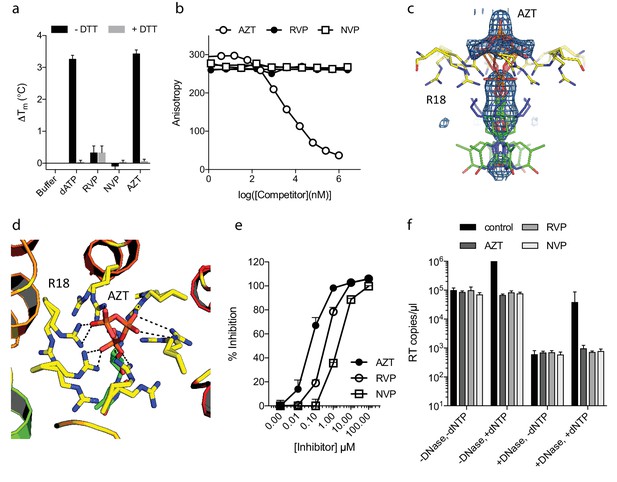
NNRTIs and NRTIs are equally potent ERT inhibitors despite differences in pore binding.
(a) Changes in hexamer capsid stability upon incubation with dATP or inhibitors in the presence and absence of DTT. Data are averaged from three replicates and representative of three independent experiments. (b) Fluorescence anisotropy competition binding experiments to hexamer capsid with inhibitors. A decrease in anisotropy indicates the ligand has displaced bound BODIPY-ATP. Averaged data from three measurements is shown and is representative of three independent experiments. (c) Electron density for NRTI AZT in complex with HIV capsid hexamer. (d) Putative hydrogen bonds between AZT and capsid reside R18. R18 adopts multiple side-chain conformations. (e) In vitro inhibition of reverse transcriptase enzyme. Data are mean of three replicates ± SD. (f) ERT experiments in presence of inhibitors at in vitro IC90 concentrations. Data are mean of three replicates ± SD.
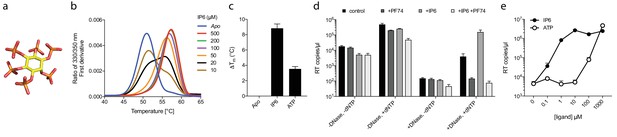
Assembly cofactor IP6 promotes ERT.
(a) IP6 molecule, indicating the non-planar phosphate arrangement. (b) IP6 stabilises capsid hexamers with a 1:1 stoichiometry. Data are representative of three different experiments. (c) IP6 makes capsid hexamers more stable than ATP. The mean of three measurements ± SD is shown and is representative of three independent experiments. (d) IP6 (100 µM) increases RT inside capsid cores (+DNase) but not RT in bulk solution (-DNase). PF74 (30 µM) prevents ERT and counteracts IP6. (e) IP6 stimulates ERT more potently than ATP. ERT data are plotted as the mean ± SD of three replicates.
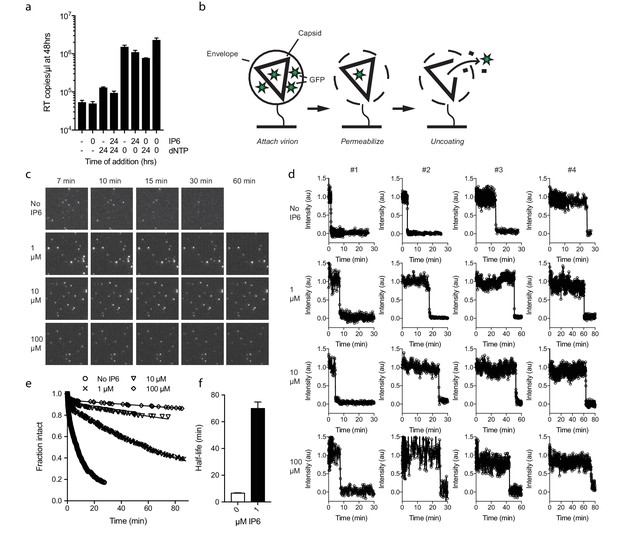
IP6 stabilises the HIV capsid both before and during DNA synthesis.
(a) IP6, dNTP, both or neither were added to capsid cores at the indicated time points and ERT measured at 48 hr in the absence of nuclease. IP6 added at 0 hr maintains HIV capsid integrity sufficiently to allow ERT when dNTPs are added 24rs later. Data are plotted as the mean ± SD of three replicates. (b) Schematic of the core opening assay. (c) Field-of-view of GFP +intact capsids at different time points and concentrations of IP6 (d) Example traces of single particle GFP release upon core opening. Traces from four individual virions undergoing core opening at different times are shown for each condition. (e) Fraction of intact cores as a function of time (f) Half-life of core opening.
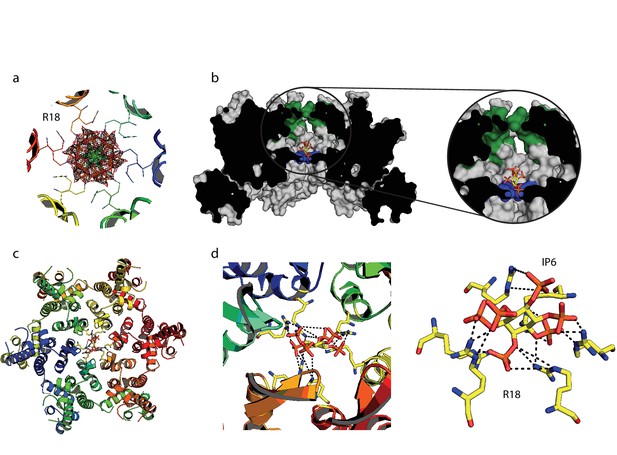
IP6 binds the HIV capsid and coordinates all R18 pore guanidino groups.
(a) View of IP6:hexamer complex down the 6-fold symmetry axis. Secondary structure is colored by monomer. R18 in each monomer adopts two alternate side-chain conformers. All symmetrically equivalent orientations of IP6 present in the structure are shown, together with the 2Fo-Fc density (mesh) centred on the ligand and contoured at 1σ. (b) View through the centre of the IP6:hexamer complex orthogonal to the 6-fold axis. The molecular surface of the capsid is shown together with a single bound IP6 molecule. N-terminal β-hairpin residues are shown in green and the R18 ring is shown in blue. (c) View down the 6-fold axis showing IP6 bound within the capsid hexamer. (d) R18 coordination by a single bound IP6 molecule. Putative interactions (2.2–4.2 Å) between each arginine and phosphate is indicated by a black dotted line.
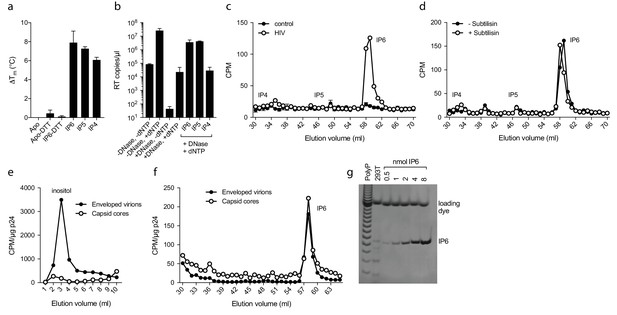
IP6 is specifically incorporated into the capsid of HIV virions.
(a) Hexamer stability in presence of 0.2 µM IP4 – IP6. Data are averaged from three replicates with SD and are representative of at least three independent experiments. (b) IP5 (1 µM) and IP6 (1 µM) promote RT accumulation in presence of nuclease. Data are plotted as the mean of three replicates with SD. (c) HIV produced in 3H-inositol-treated cells, acid-extracted and Sax-HPLC fractioned reveals specific packaging of IP6 but not IP4 or IP5 into virions. Supernatant of mock transfected cells processed in parallel with viral samples was used as a control. Scintillation data are single measurements representative of three independent experiments. (d) IP6 incorporation in virions treated with subtilisin to remove microvesicle contamination (Figure 7—figure supplement 1). (e) Comparison of inositol levels in samples of intact enveloped virions or purified capsid cores (measured as CPM per µg of p24 capsid). (f) As in (e) but comparing the levels of IP6. Enveloped virions and capsid cores have equal IP6 incorporation. (g) Toluidine blue PAGE of IP6 purified from 293 T cells with known IP6 and polyphosphate (PolyP) standards.
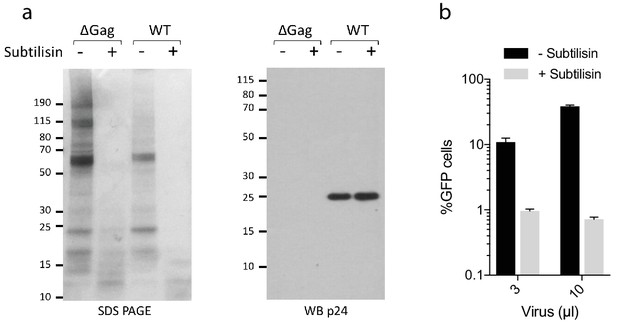
Subtilisin treatment of HIV virions.
Subtilisin treatment of HIV virions or control supernatant of cells transfected without gag encoding plasmid. (a) Samples were run on SDS PAGE and also blotted for p24 protein. (b) Effect of subtilisin treatment on HIV-1 infection.
Tables
Data collection and refinement statistics
https://doi.org/10.7554/eLife.35335.004| 6ERM | 6ERN | 6ES8 | |
|---|---|---|---|
| Data collection | |||
| Space group | P6 | P6 | P6 |
| Cell dimensions | |||
| a, b, c (Å) | 91.04, 91.04, 56.53 | 90.85, 90.85, 56.75 | 91.30, 91.30, 57.20 |
| a, b, g (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 78.85–2.00 (2.03–2.00) | 78.68–2.36 (2.44–2.36) | 79.08–1.90 (2.0–1.9) |
| Rmeas | 13.3 (59.0) | 7.0 (28.7) | 8.4 (87.8) |
| CC1/2 (%) | 99.0 (90.9) | 99.8 (95.0) | 99.1 (94.3) |
| I / σI | 10.8 (6.1) | 15.4 (3.9) | 10.8 (1.6) |
| Completeness (%) | 100.0 (100.0) | 92.6 (92.6) | 91.5 (94.1) |
| Redundancy | 9.3 (9.2) | 4.9 (4.7) | 4.6 (4.4) |
| Resolution (Å) | 2.0 | 2.36 | 1.90 |
| No. reflections | 18217 | 11142 | 21569 |
| Rwork/Rfree | 0.23/0.18 | 0.24/0.19 | 0.22/0.18 |
| No. of atoms | 1808 | 1674 | 1816 |
| Protein | 1569 | 1562 | 1623 |
| Ligand/ion | 30 | 30 | 71 |
| Water | 209 | 82 | 122 |
| B-factors | |||
| Protein | 29.0 | 36.87 | 33.2 |
| Ligand/ion | 71.6 | 77.49 | 60.2 |
| Water | 36.3 | 38.39 | 42.7 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.02 | 0.017 | 0.02 |
| Bond angles (°) | 1.90 | 1.82 | 1.90 |
-
*Values in parentheses are for highest-resolution shell.
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35335.012





