N-glycosylation in the protease domain of trypsin-like serine proteases mediates calnexin-assisted protein folding
Figures
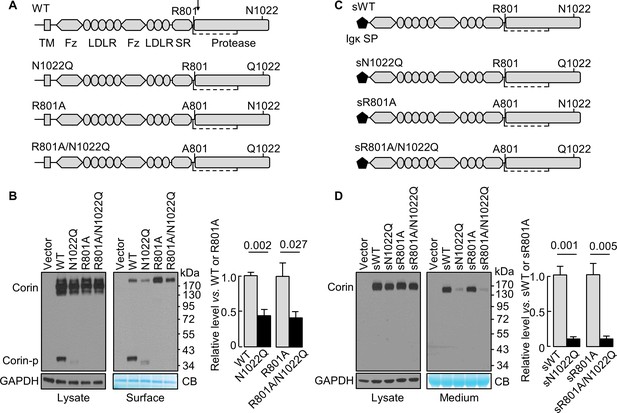
N-glycosylation at N1022 in single-chain and soluble corin.
(A) Illustration of human corin WT and mutants with or without R801 activation site and N1022 N-glycosylation site. TM: transmembrane; Fz: frizzled; LDLR: LDL receptor; SR: scavenger receptor. An arrow indicates the PCSK6-mediated activation cleavage site at Arg801 (R801). A disulfide bond linking the pro-peptide region and the protease domain is indicated by a dashed line. (B) Western blotting, under reducing conditions, of corin proteins in lysates (left) or on the cell surface (right) from HEK293 cells. Corin zymogen bands (Corin) and the cleaved protease domain fragment (Corin-p) are indicated. Levels of GAPDH in cell lysates and a Coomassie Blue (CB)-stained non-specific protein in biotin-labeled cell surface proteins were used to assess amounts of proteins in each sample. Relative corin levels on the cell surface are estimated by densitometric analysis of western blots. Data are means ± S.E. from four independent experiments. p-Values are shown in the bar graph. (C) Illustration of soluble corin (sWT) and mutants, in which the cytoplasmic and transmembrane domains were replaced by the Igκ signal peptide (SP). (D) Western blotting of soluble corin in lysates (left) and medium (right) from HEK293 cells. Levels of GAPDH in cell lysates and a Coomassie Blue (CB)-stained non-specific protein in the conditioned media were used to assess amounts of proteins in each sample. Relative levels of the secreted corin in the medium were estimated by densitometric analysis of Western blots. Data are means ± S.E. from three independent experiments. p-Values are shown in the bar graph.
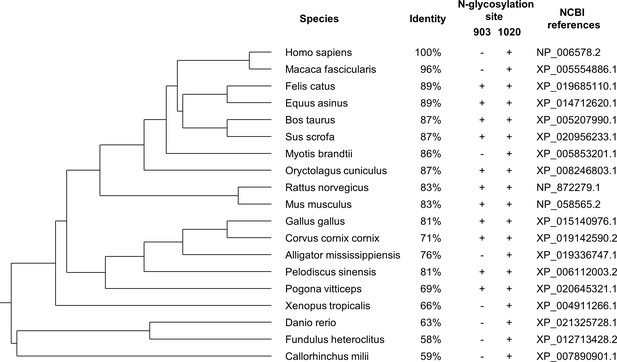
The phylogenetic tree of corin proteins in different species.
The phylogenetic relationships of corin in different species were evaluated using the COBALT server at the National Center for Biotechnology Information based on the full-length corin amino acid sequences. The identity (vs. Homo sapiens), the presence of N-glycosylation sites (corresponding to H. sapiens) in the protease domain, and the NCBI references are shown. The N-glycosylation site at N1022 is conserved in all species, whereas the N-glycosylation site corresponding to residue 903 in H. sapiens is less conserved. (+) presence; (-) absence.

Alignments of the protease domain of trypsin-like serine proteases.
The amino acid sequences were aligned using the COBALT server. The N-glycosylation sites (red) are indicated. Corin has one N-glycosylation site that is also present in testisin; enteropeptidase (EK) has four N-glycosylation sites that are also present in plasma kallikrein (PKK), testisin or factor VII (FVII); and prothrombin (PT) has one N-glycosylation site that is also present in tryptase γ.
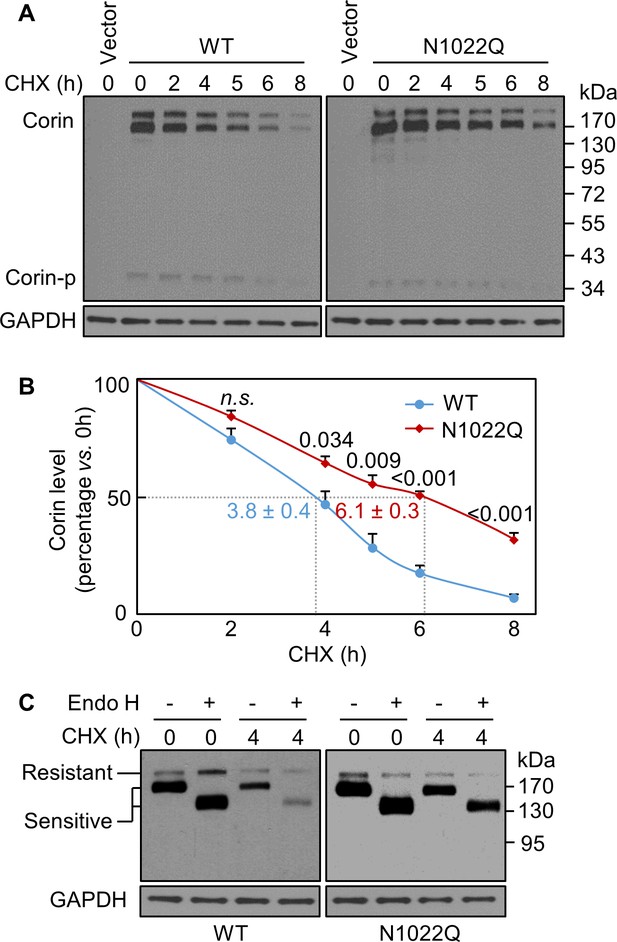
Analysis of intracellular corin by CHX-based protein chase and Endo H digestion.
(A) Western blotting of corin in HEK293 cells treated without (0) or with CHX over time (h). (B) Percentages of corin WT and the mutant N1022Q levels, with corresponding levels at 0 hr being 100%, were estimated by densitometric analysis of Western blots. In addition to corin zymogen bands (Corin), a weak Corin-p band was detected, which likely represented activated corin on the cell surface. Data are means ± S.E. from four independent experiments. P values vs. WT at the same time point are shown. n.s.: not significant. The half-lives in h for WT (blue) and N1022Q (red) are indicated. (C) Endo H digestion of proteins from HEK293 cells without (0) or with CHX treatment for 4 hr. Corin proteins without (-) or with (+) Endo H digestion were analyzed by western blotting. Endo H-sensitive and resistant bands are indicated.
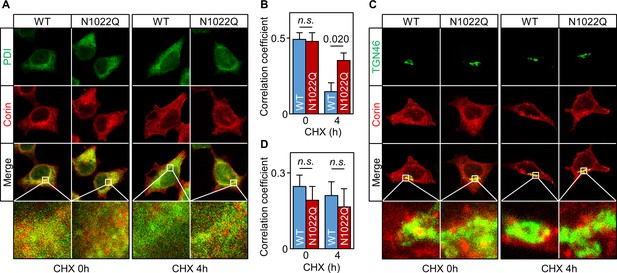
Intracellular distribution of corin WT and the N1022Q mutant.
(A) Co-staining of corin and PDI in HEK293 cells expressing WT corin and the N1022Q mutant without (0) or with CHX treatment for 4 hr. (B) Correlation of red (corin) and green (PDI) colors within individual cells was analyzed by Pearson’s correlation coefficient. p-Value is shown. n.s.: not significant. (C) Co-staining of corin and TGN46 (green) in HEK293 cells expressing WT corin and the N1022 mutant without (0) or with CHX treatment for 4 hr. (D) Correlation of red (corin) and TGN46 (green) colors within individual cells was analyzed by Pearson’s correlation coefficient. Data are means ± S.E. from five independent experiments.
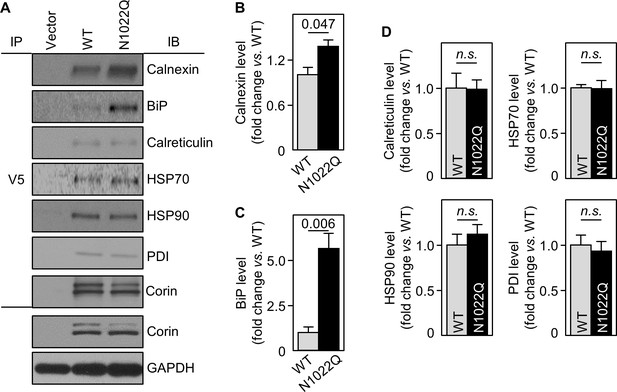
Interactions between corin and ER chaperones.
(A) Corin proteins in HEK293 cells expressing WT corin and the N1022Q mutant were immunoprecipitated (IP) with an anti-V5 antibody that recognizes the C-terminal V5 tag in corin. Chaperones that co-precipitated with corin were analyzed by immunoblotting (IB, top six panels). Corin in V5 pull-down samples was verified. Corin and GAPDH in the cell lysates were analyzed as additional controls (bottom two panels). Relative levels of calnexin (B), BiP (C), calreticulin, HSP70, HSP90 and PDI (D) were estimated by densitometric analysis of western blots. Data are means ± S.E. from three independent experiments. p-Values are shown in bar graphs. n.s.: not significant.
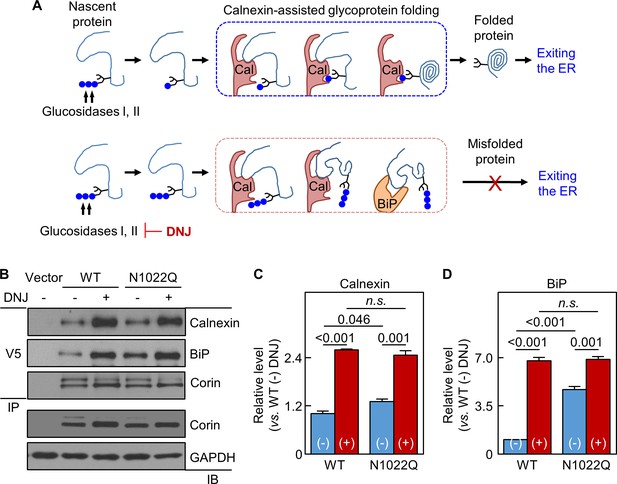
Analysis of calnexin interaction.
(A) A model of calnexin-assisted glycoprotein folding. Cal: calnexin; blue dots: glucose residues. DNJ inhibits glucosidases I and II. (B) Co-immunoprecipitation (IP) and western blotting (IB) of corin associated calnexin and BiP in HEK293 cells expressing WT corin or the N1022Q mutant without (-) or with (+) DNJ treatment (top two panels). Corin in V5 pull-down samples was verified (third panel). Corin and GAPDH in the lysates were also verified (bottom two panels). Relative calnexin (C) and BiP (D) levels were estimated by densitometric analysis of western blots. Data are means ± S.E. from three independent experiments. p-Values are shown in bar graphs. n.s.: not significant.
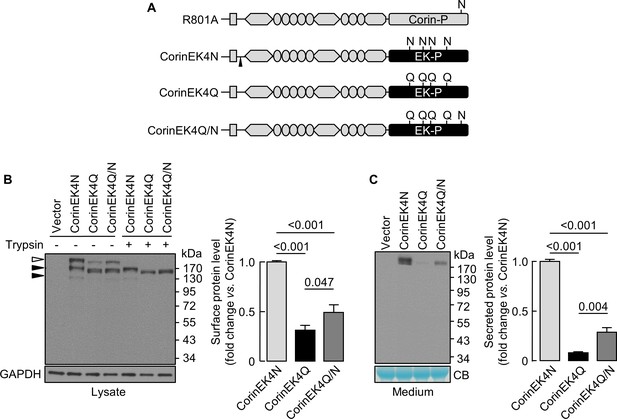
Analysis of N-glycosylation in the protease domain of corin-EK chimeras.
(A) In CorinEK4N, the corin protease domain (Corin-P) was replaced by the EK protease domain (EK-P). The ADAM10-mediated shedding site is indicated by an arrowhead. In CorinEK4Q, all four N-glycosylation sites in the EK protease domain were mutated by Gln (Q) residues. In CorinEK4Q/N, a new N-glycosylation site corresponding to N1022 in corin was added to CorinEK4Q. (B) Western blotting of CorinEK4N, CorinEK4Q and CorinEK4Q/N in transfected cells treated without (-) or with (+) trypsin before the cells were lysed. GAPDH levels in cell lysates were used to assess amounts of proteins in each sample. (C) Western blotting of shed corin fragments the in medium. Corin levels on the cell surface (B) and in the medium (C) were estimated by densitometric analysis of western blots. In (C), levels of a Coomassie Blue (CB)-stained non-specific protein were used to assess amounts of proteins in each sample. Data are means ± S.E. from at least three independent experiments. p-Values are shown in bar graphs.
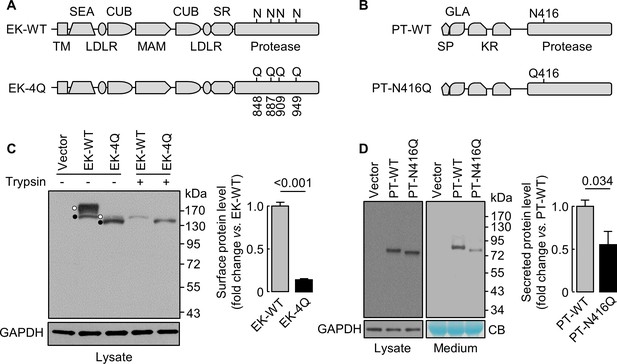
Analysis of N-glycosylation sites in the protease domain of EK and prothrombin.
(A) Illustration of EK WT and the mutant lacking the indicated N-glycosylation sites (EK-4Q). EK domains include transmembrane (TM), SEA, LDLR, CUB, MAM, scavenger receptor (SR) and protease domains. (B) Illustration of prothrombin WT and the PT-N416Q mutant lacking the N-glycosylation site in the protease domain. Prothrombin domains include signal peptide (SP), Gla (GLA), kringle (KR) and protease domains. (C) Western blotting of EK-WT and EK-4Q in HEK293 cells without (-) or with (+) trypsin treatment before the cells were lysed. The cell surface (trypsin-sensitive; white dots) and intracellular (trypsin-resistant; black dots) bands are indicated. Relative levels of surface EK bands in EK-WT and EK-4Q were estimated by densitometric analysis of western blots. Data are means ± S.E. from four independent experiments. p-Value is shown. (D) Western blotting of PT-WT and PT-N416Q in cell lysates (left) and the medium (right) from HEK293 cells. Relative levels of PT-WT and PT-N416Q in the medium were estimated by densitometric analysis of western blots. Levels of GAPDH in cell lysates and a Coomassie Blue (CB)-stained non-specific protein in the conditioned medium were used to assess protein amounts in each sample. Data are means ± S.E. from four independent experiments. p-Value is shown.
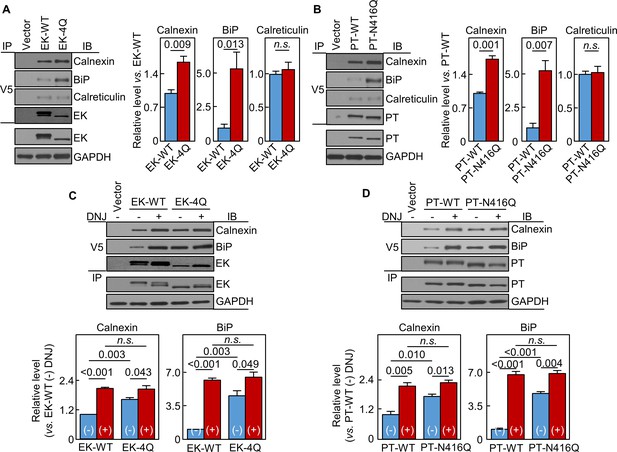
Interactions of EK and prothrombin with chaperones.
Co-immunoprecipitation (IP) and Western blotting (IB) of EK-WT and EK-4Q (A) or PT-WT and PT-N416Q (B) binding to calnexin, BiP and calreticulin (top three panels). EK (A) and PT (B) proteins in V5 pull-down samples were verified. EK, PT and GAPDH in cell lysates were also verified by western blotting (bottom two panels). Relative levels of calnexin, BiP and calreticulin associated with EK-WT and EK-4Q (A) or PT-WT and PT-N416Q (B) were estimated by densitometric analysis of Western blots. Data are means ± S.E. from three and four independent experiments, respectively. p-Values are shown in bar graphs. IP and IB analysis of EK-WT and EK-4Q (C) or PT-WT and PT-N416Q (D) binding to calnexin and BiP in the cells without (-) or with (+) DNJ treatment (top two panels). EK (A) and PT (B) proteins in V5 pull-down samples were verified. EK, PT and GAPDH proteins in cell lysates were also verified. Relative calnexin and BiP levels associated with EK-WT and EK-4Q (C) or PT-WT and PT-N416Q (D) were estimated by densitometric analysis of Western blots. Data are means ± S.E. from three independent experiments. p-Values are shown in bar graphs.
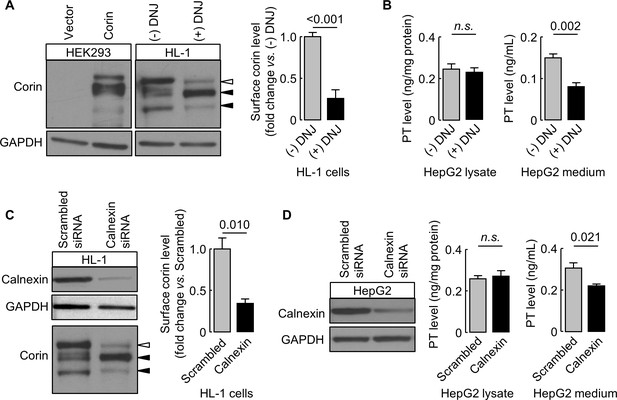
Effects of DNJ treatment and calnexin knockdown.
HL-1 (A) and HepG2 (B) cells were cultured without (-) or with (+) DNJ. Recombinant human corin expression in transfected HEK293 cells were included as a control (A, left). Corin cell surface expression (A) and prothrombin expression in cell lysates and secretion in the medium (B) were analyzed by western blotting and ELISA, respectively. Levels of corin cell surface band (open arrowhead) were estimated by densitometric analysis of western blots. Data are means ± S.E. from four independent experiments. p-Values are shown in bar graphs. To knockdown calnexin expression, HL-1 (C) and HepG2 (D) cells were transfected with calnexin-targeting or control scrambled siRNAs. Calnexin expression levels in the transfected cells were verified by western blotting. Corin cell surface expression (C) and prothrombin expression in cell lysates and secretion in the medium (D) were analyzed by western blotting and ELISA, respectively. Data are means ± S.E. from three independent experiments. p-Values are shown in bar graphs. n.s.: not significant.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | Corin | NCBI | NM_006587.3 | |
| Gene (H. sapiens) | Prothrombin, PT | NCBI | NM_002772.2 | |
| Gene (H. sapiens) | Enteropeptidase, EK | NCBI | NM_000506.4 | |
| Genetic reagent (H. sapiens) | Calnexin (siRNA kit) | Origene | SR300576 | |
| Genetic reagent (Mus musculus) | Calnexin (siRNA kit) | Origene | SR417891 | |
| Cell line (H. sapiens) | HEK293 | ATCC | CRL-1573 | STR profiling, no mycoplasma contamination |
| Cell line (M. musculus) | HL-1 | PMID: 21518754, EMD Millipore: SCC065 | From Dr. William Claycomb | No mycoplasma contamination |
| Cell line (H. sapiens) | HepG2 | ATCC | HB-8065 | STR profiling, no mycoplasma contamination |
| Transfected construct (H. sapiens) | Corin plasmid | PMID: 14559895 | ||
| Transfected construct (H. sapiens) | sCorin plasmid | This paper | ||
| Transfected construct (H. sapiens) | CorinEK plasmid | This paper | ||
| Transfected construct (H. sapiens) | EK plasmid | This paper | ||
| Transfected construct (H. sapiens) | PT plasmid | This paper | ||
| Antibody | Anti-V5 | Thermo Fisher | R96025 | |
| Antibody | Anti-V5-HRP | Thermo Fisher | R96125 | |
| Antibody | Anti-GAPDH | EMD Millipore | MAB374 | |
| Antibody | Anti-PDI | Abcam | ab3672 | Immunostaining |
| Antibody | Anti-TGN46 | Abcam | ab50595 | |
| Antibody | Anti-Igg (mouse)-Alexa-594 | Thermo Fisher | A-21203 | |
| Antibody | Anti-Igg (rabbit)-Alexa-488 | Thermo Fisher | A-11008 | |
| Antibody | Anti-calnexin (human) | Cell Signaling | 2679T | |
| Antibody | Anti-BiP | Cell Signaling | 3177T | |
| Antibody | Anti-calreticulin | Cell Signaling | 12238S | |
| Antibody | Anti-HSP70 | Cell Signaling | 4872T | |
| Antibody | Anti-HSP90 | Cell Signaling | 4877T | |
| Antibody | Anti-PDI | Cell Signaling | 3501T | Western blotting |
| Antibody | Anti-Igg (mouse)-HRP | KPL | 474–1806 | |
| Antibody | Anti-Igg (rabbit)-HRP | KPL | 474–1516 | |
| Antibody | Anti-calnexin (mouse) | Abcam | ab75125 | |
| Antibody | Anti-corin (mouse) | Homemade | PMID: 26259032 | |
| Recombinant DNA reagent | pSecTag/FRT/V5-His Expression kit (vector) | Thermo Fisher | K602501 | |
| Recombinant DNA reagent | pcDNA 3.1/V5-His Expression kit (vector) | Thermo Fisher | K480001 | |
| Commercial assay or kit | ELISA kit (prothrombin) | Abcam | ab108909 | |
| Chemical compound, drug | 1-deoxynojirimycin, DNJ | Alfa Aesar | J62602-MC |
Additional files
-
Supplementary file 1
Proteins that differentially bound to WT corin and the N1022Q mutant identified in proteomic analysis.
- https://doi.org/10.7554/eLife.35672.013
-
Supplementary file 2
Proteins with a ratio of ≥ 2 fold between WT corin and the N1022Q mutant.
- https://doi.org/10.7554/eLife.35672.014
-
Supplementary file 3
Information of the DNA inserts in the expression plasmids used in this study.
- https://doi.org/10.7554/eLife.35672.015
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35672.016





