Coalescing beneficial host and deleterious antiparasitic actions as an antischistosomal strategy
Figures
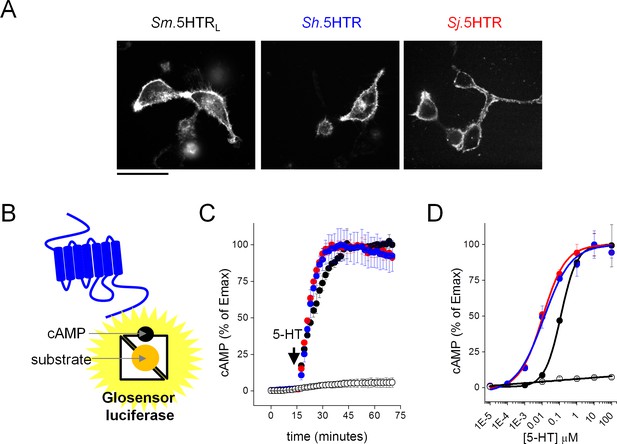
Functional expression of Schistosoma serotonin receptors.
(A) Confocal images showing cell surface expression in mammalian HEK293 cells of 5-HT receptors cloned from S. mansoni (Sm.5HTRL), S. haematobium (Sh.5HTR) and S. japonicum (Sj.5HTR) localized by COOH-terminally tagged eGFP. Scalebar, 50 µm. (B) Schematic of luminescent cAMP sensor bioassay. cAMP generated by schistosome 5-HTRs (blue) binds the engineered GloSensor luciferase, switching the sensor to a more active conformation resulting in enhanced luminescence output. (C) Kinetics of signal following the addition of 5-HT (1 µM) to HEK293 cells co-transfected with luminescent cAMP sensor and individual schistosome 5-HT receptors. Open circles, cells not transfected with 5-HT receptor, colored circles represent measurements in cells transfected with Sm.5HTRL (black), (Sh.5HTR (blue) and Sj.5HTR (red). (D) Serotonin dose-response curves for each of the three receptors (colored circles) and HEK293 cells expressing the cAMP sensor alone (open circles). Data reflect mean ± standard error of at least 3 biological replicates.

Schistosome 5-HT receptor protein alignment.
Predicted sequences from S. japonicum and S. haematobium (545 and 547 amino acids long) closely match the ‘long’ isoform of the previously cloned S. mansoni receptor (Sm.5HTRL, 547 amino acids; NCBI accession # KX150867, [Chan et al., 2017]). No sequences matching the previously reported ‘short’ isoform (Sm.5HTRL, 463 amino acids; NCBI accession # KF444051 [Patocka et al., 2014]) were identified in S. mansoni, S. japonicum or S. haematobium. Regions delimiting the N-terminus and third intracellular loop shown in red. Alignment with human HTR7 (UniProt entry P34969) is shown.
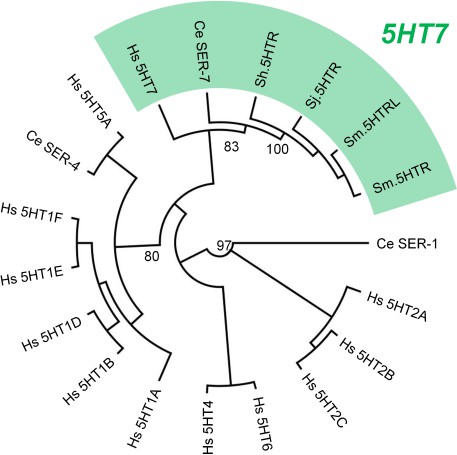
Cladogram of schistosome 5-HT receptors.
Schistosome 5.HTR sequences cluster with known 5-HT7 receptors (human 5HTR7, C. elegans SER-7). Sequences were aligned with MUSCLE and maximum likelihood phylogeny computed with PhyML(v3.1), 500 bootstrap replicates.

Performance of the Sm.5HTRL luminescent cAMP reporter assay.
(A) Coefficient of variation (CV%) for signal in response to increasing concentrations of 5-HT (white = 0.1 µM, gray = 1 µM, black = 10 µM) relative to basal levels (no 5-HT addition). Dotted line represents threshold for acceptable limit of 15%. CV%=(σ5-HT/µ5-HT) x100. (B) Signal window (SW) for serotonin response relative to basal levels. Dotted line = acceptable limit of 4-fold. SW = (µ5-HT - µbasal - 3(σ5-HT + σbasal)) / σ5-HT. (C) Z’ factor for signal evoked by increasing concentrations of serotonin. Dotted line represents acceptable threshold of 0.5. Z’ factor = 1 - ((3σ5-HT + 3σbasal) / |µ5-HT - µbasal|). (D) Z factor for control antagonist (bromocriptine, BRM) inhibition of 5-HT response. Z factor = 1 - ((3σ5-HT + 3σ5-HT+BRM) / |µ5-HT - µ5-HT+BRM|). (A–D) All parameters were calculated from 48 wells per concentration of compound in a 384 well plate.
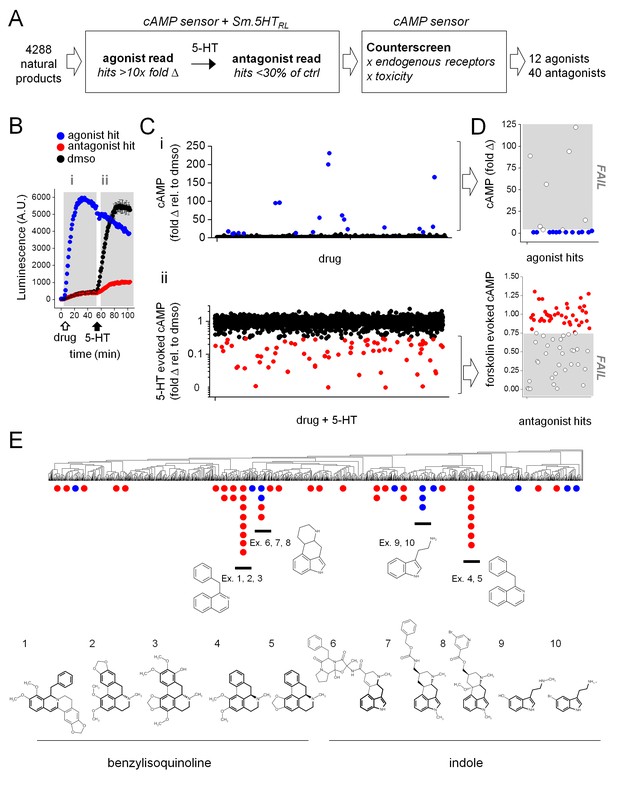
High-throughput screen of natural product libraries against Sm.5HTRL.
(A) Assay workflow for drug screen. Compounds were screened against HEK293 cells stably expressing a cAMP sensor and Sm.5HTRL, followed by counter-screening hits against a cell line lacking Sm.5HTRL. (B) Kinetic readout of luminescence from cAMP biosensor following addition of (i) test compound (10 µM, open arrow; agonist hits increase luminescence), followed by (ii) the addition of 5-HT (1 µM, solid arrow; antagonist hits decrease luminescence relative to controls). (C) Scatter plot of compounds assaying for (i) agonists and (ii) antagonists. Putative hits surpassing threshold are shown in color (blue, agonists; red, antagonists). (D) Counter-screen of putative hit compounds from (C) against cell lines lacking 5-HT receptor to exclude compounds with off-target increases in cAMP (top) or compounds that reduce forskolin (25 µM) evoked cAMP signals (bottom). (E) Hits clustered into compound classes by structure. Groups contain common ring systems such as benzylisoquinoline (ex. 1. ST059293, 2. Nantenine, 3. 785163, 4. Nuciferine, 5. Remerine) or indole (6. Ergotamine, 7. Metergoline, 8. Nicergoline, 9. N-methylserotonin, 10. 5-bromotryptamine) structures. Blue = agonists, red = antagonists.
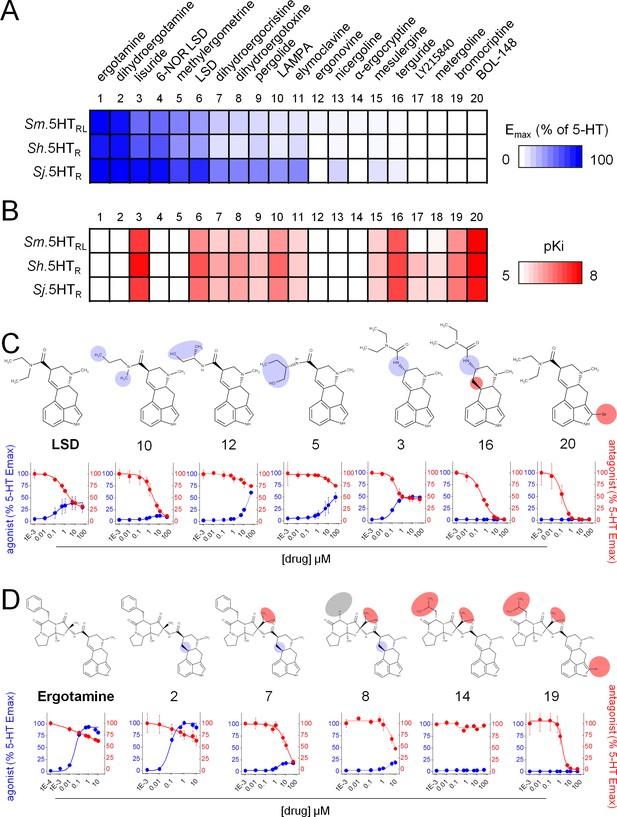
Structure activity relationship of ergot alkaloid compounds.
Ergot alkaloids display a spectrum of activity against Schistosoma 5-HT receptors ranging from (A) agonists (expressed as maximal cAMP response relative to 5-HT) to (B) antagonists (estimated receptor affinity expressed as pKi). (C) Structure activity relationship within a lysergic acid amide series. Derivatives of the partial agonist LSD displayed modified activity following modification of the diethylamide moieties (Danso-Appiah and De Vlas, 2002; Fallon and Doenhoff, 1994; Cheever, 1969; Wilson, 2009) or the ergoline ring system (Geary et al., 1992; Chan et al., 2016c). (D) Structure activity relationship within a ergopeptine chemical series. Derivatives of the full agonist ergotamine with altered activity following modification of the tripeptide structure (Barlow and Meleney, 1949; Andrews et al., 1983; Ismail et al., 1996) or the ergoline ring system (Chan et al., 2016b). Compound numbering reflects rank order of efficacy across the schistosome 5-HTRs: 1. Ergotamine; 2. Dihydroergotamine; 3. Lisuride; 4. 6-NOR LSD; 5. Methylergometrine; 6. LSD; 7. Dihydroergocristine; 8. Dihydroergotoxine; 9. Pergolide; 10. LAMPA; 11. Elmoclavine; 12. Ergonovine; 13. Nicergoline; 14. α-ergocryptine; 15. Mesulergine; 16. Terguride; 17. LY215840; 18. Metergoline; 19. Bromocriptine; 20. BOL-148. Data represent mean ± standard error of at least three biological replicates.
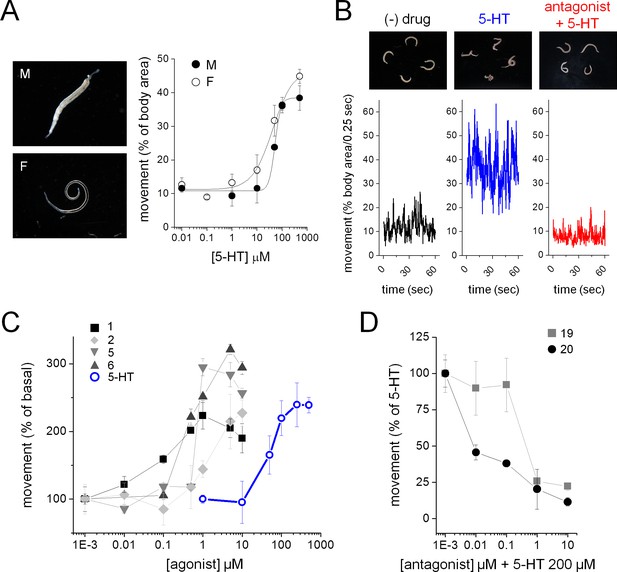
Ergot alkaloid action against adult S. mansoni parasites ex vivo.
(A) Serotonin induced increase in movement of S. mansoni adult male (M, solid circles) and female (F, open circles) parasites. (B) Adult S. mansoni movement in the absence of drug (black trace), following 5-HT addition (200 µM, blue), or exposure to a Sm.5HTRL antagonist (BOL-148, 1 µM) and subsequent 5-HT (200 µM, red) addition. (C) Dose-response curves for Sm.5HTRL agonist evoked stimulation of adult schistosome movement. Ergot alkaloids = solid symbols, 5-HT = open blue circles. (D) Dose-response curves for Sm.5HTRL antagonist inhibition of 5-HT (200 µM) evoked movement. Worm movement data (A, C–D) reflect mean ± standard error of the mean for at least 3 biological replicates.
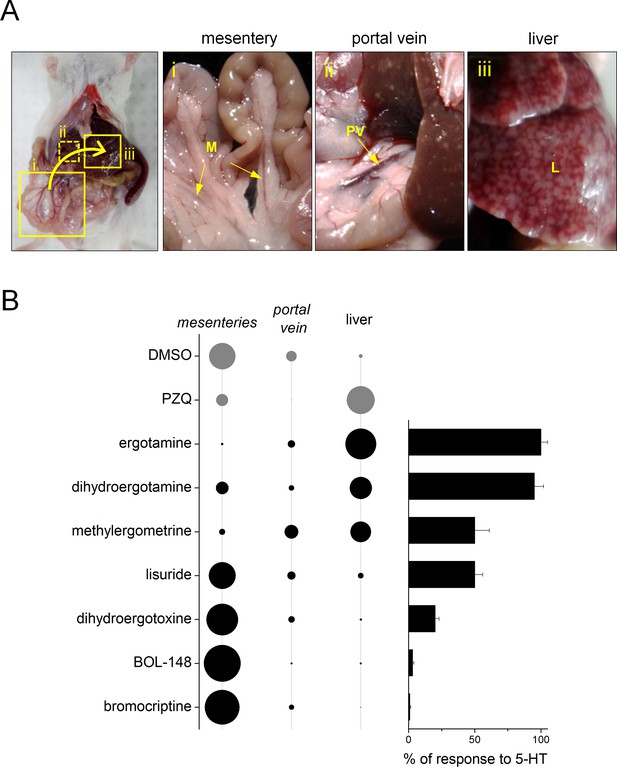
Ergot alkaloid agonists evoke an acute hepatic shift of parasites in vivo.
(A) Location of mouse vasculature dissected after drug treatment to resolve worm location. Adult parasites normally reside in the mesenteric vasculature (i), but following anthelmintic treatment shift to the portal vein (ii) and liver (iii). (B) Left, quantification of percent worm burden found in either the mesenteries, portal vein or liver following treatment with DMSO (negative control), praziquantel (100 mg/kg, positive control) and various ergot alkaloid compounds. Diameter of circle represents proportionality of distribution. Right, bar chart represents efficacy of the same series of compounds at Sm.5HTRL.
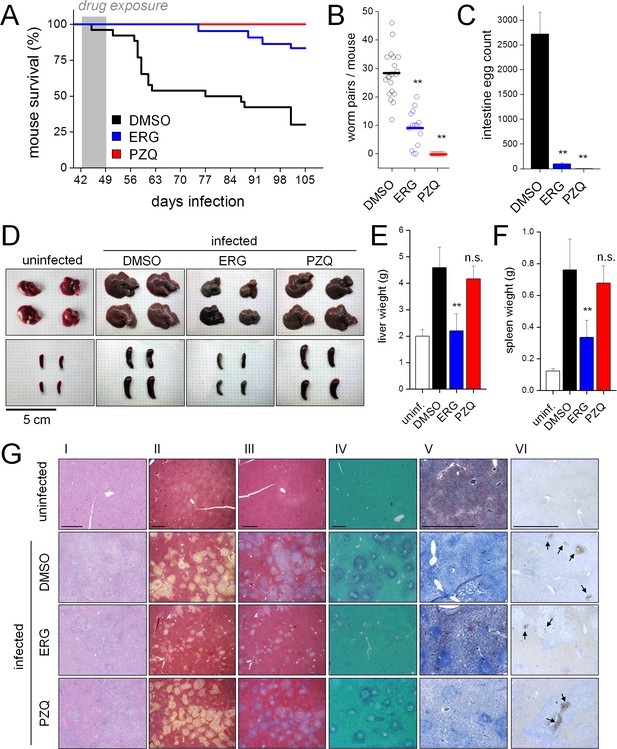
Ergotamine resolves infections and mitigates pathology of schistosomiasis in vivo.
(A) Survival of mice infected with S. mansoni and treated for one week (days 42–49 post infection) with either vehicle control (DMSO. n = 26 mice), ergotamine (ERG, 60 mg/kg twice daily. n = 22 mice) or praziquantel (PZQ, 50 mg/kg daily. n = 10 mice). (B) Worm burden of mice treated as in A and sacrificed 49 days post-infection. (C) Intestinal egg counts of mice sacrificed in B. (D) Representative livers and spleens from uninfected mice and infected mice exposed to vehicle (DMSO), ergotamine (ERG) or praziquantel (PZQ). Quantification of hepatomegaly (E) and splenomegaly (F) in uninfected mice (open columns) and infected mice following drug treatment (solid columns). (G) Histology of liver sections control uninfected and infected (DMSO) mice, as well as drug treated (ERG and PZQ) infected animals (scale = 1 mm). Liver histology using (I) H and E staining, (II) Movat’s stain (collagen, yellow), (III) Masson’s trichrome stain (collagen, blue), (IV) aldehyde fuchsin stain (elastin, purple), (V) Oil red O stain (lipid, red), and (VI) caspase-3 activation (brown puncta, arrows). Higher magnification images of egg granulomas for each staining condition are shown in Figure 6—figure supplement 2.
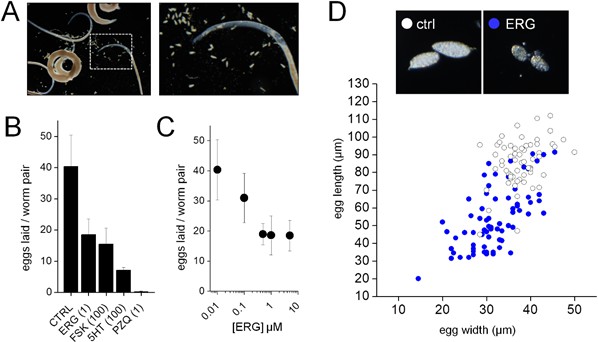
Ergotamine inhibition of S. mansoni egg laying during ex vivo culture.
(A) Eggs laid by adult S. mansoni cultured ex vivo as male and female pairs. (B) Average number of eggs laid per worm pair over the course of five days for worms treated with DMSO (0.1% v/v), ergotamine (ERG, 1 µM), forskolin (FSK, 100 µM), serotonin (5-HT, 100 µM) or praziquantel (PZQ, 1 µM). (C) Dose response for ergotamine inhibition of egg laying. (D) Morphological changes in eggs laid by ergotamine (1 µM, blue circles) treated worms relative to control (open circles). Data reflects mean ± standard error of at least 3 biological replicates.

Liver egg granulomas in control and drug treated mice.
Histology of sections of liver from uninfected mice and infected mice treated with DMSO, ergotamine (ERG, 60 mg/kg), praziquantel (PZQ, 50 mg/kg). (I) H and E staining, (II) Movat’s stain (collagen, yellow), (III) Masson’s trichrome stain (collagen, blue), (IV) aldehyde fuchsin stain (elastin, purple), (V) Oil red O stain (lipid, red), and (VI) caspase-3 activation (brown puncta). Scale = 500 µm.
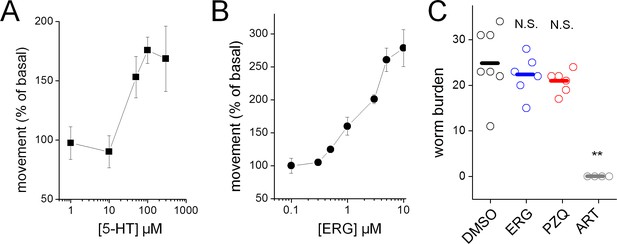
Effects of ergotamine on immature S. mansoni.
Schistosomes were harvested from mice 4 weeks post infection and the movement of male worms was assayed in response to (A) serotonin (5-HT) and (B) ergotamine (ERG). (C) Effect of ergotamine (ERG, 60 mg/kg) and the anthelmintics praziquantel (PZQ, 50 mg/kg) and artemether (ART, 100 mg/kg) on schistosome infection in vivo. Drugs were administered to mice between weeks 3–4 post-infection, and animals were euthanized at 7 weeks post-infection to assess worm burden. Open circles = worm burdens of individual mice, bars = mean values. N.S. = not significant. **p<0.001.

Lack of hepatic microvesicular steatosis in drug treated livers.
Histological H and E staining of formalin fixed, paraffin embedded liver sections from uninfected mice and infected mice treated with DMSO, ergotamine (ERG, 60 mg/kg), praziquantel (PZQ, 50 mg/kg). Scale = 50 µm.
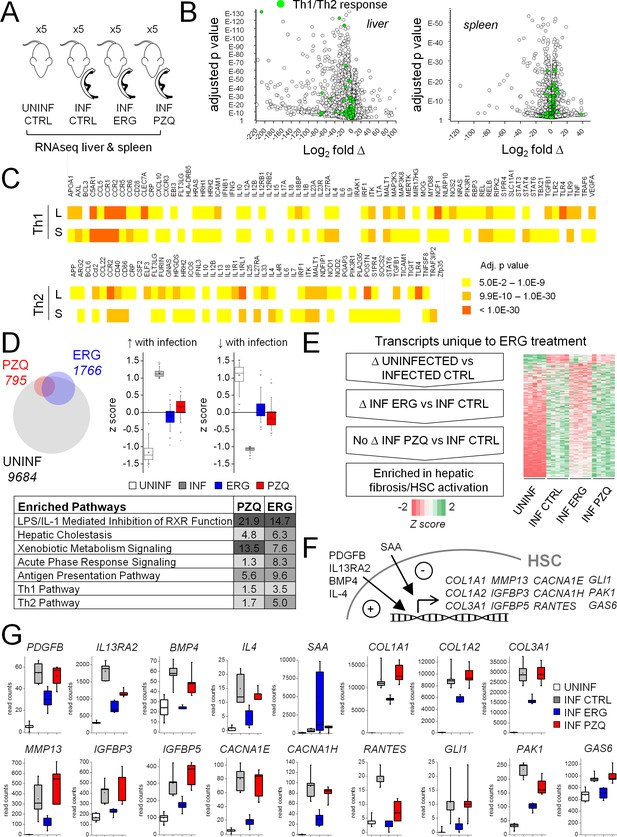
Effect of ergotamine on immune response and hepatic stellate cell activation.
(A) Schematic of schistosome infection and drug treatment cohorts analyzed. Livers and spleens were harvested from infected mice treated with vehicle control, ergotamine (ERG 60 mg/kg, twice daily for 7 days) or praziquantel (PZQ 50 mg/kg, once daily for 5 days), as well as uninfected littermate controls. Five mice were used per cohort. (B) Volcano plots showing transcriptional changes of the liver and spleens from infected mice compared to uninfected littermates. Green shading highlights gene products involved in Th1/Th2 cell signaling. (C) Heat map of liver (L) and spleen (S) changes in Th1 and Th2 signaling pathways - color intensity corresponds to significance (FDR adjusted p-values). (D–G) Effects of anthelmintics on hepatic gene expression of schistosome infected mice. (D) Top left - differentially expressed transcripts comparing the livers of infected controls to uninfected littermates (grey), PZQ-treated animals (red) and ERG-treated animals (blue). Top right - expression of transcripts either up-regulated or down-regulated in infected livers relative to uninfected livers (z score ≥1 or ≤−1) in control and drug treated animals. Bottom, transcripts differentially expressed between PZQ-treated or ERG-treated and control infections represent pathways involved in liver function, drug metabolism and immune response. Ingenuity Pathway Analysis enrichment scores reflect -log10 p-value of PZQ and ERG datasets (p-value 0.05 = -log10 1.3). (E) Filtering of transcripts changing with schistosomiasis infection that are affected by ERG but not PZQ, resulting in an enrichment for gene products involved in hepatic fibrosis/HSC activation. (F) Role of various gene products in HSC activation, and (G) read counts for these transcripts across uninfected (open box), infected (grey box) and ERG-treated (blue box) and PZQ-treated (red box) cohorts. Box plots represent 25–75 percentile, whiskers 5–95 percentile. Inset; bar = median, square = mean. Read count data for liver and spleen samples available in Supplementary files 2 and 3.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line | HEK293 cells | Company | ATCC; CRL-1573 | |
| Biological sample (Schistosoma mansoni) | Schistosoma mansoni | Stock Center | BEI: NR-34792 | Biodefense and Emerging Infections Research Resources Repository (BEI Resources) |
| Biological sample (Schistosoma japonicum) | Schistosoma japonicum | Stock Center | BEI: NR-45100; BEI:NR-45101 | Biodefense and Emerging Infections Research Resources Repository (BEI Resources) |
| Biological sample (Schistosoma hematobium) | Schistosoma hematobium | Stock Center | BEI: NR-45102; BEI:NR-45103 | Biodefense and Emerging Infections Research Resources Repository (BEI Resources) |
| Recombinant DNA reagent | Sm.5HTRL | Data repository | GenBank accession KX150867 | |
| Recombinant DNA reagent | Sj.5HTR | Data repository | GenBank accession number MG813904 | |
| Recombinant DNA reagent | Sh.5HTR | Data repository | GenBank accession number MG813903 | |
| Commercial assay or kit | GloSensor 22F cAMP sensor | Company | Promega; E2301 | |
| Commercial assay or kit | D-luciferin sodium salt | Company | Gold Biotechnology; LUCK-100 | |
| Commercial assay or kit | Cleaved caspase −3 | Company | Biocare; CP-229C | |
| Chemical compound, drug | NCI Natural Products Set IV | Other | National Institutes of Health, NCI Development Therapeutics Program (DTP) | |
| Chemical compound, drug | Natural Product Library | Company | TimTec: NPL-800 | |
| Chemical compound, drug | Natural Derivatives Library | Company | TimTec: NDL-3000 | |
| Chemical compound, drug | LSD | Other | NIDA; 7315–006 | National Institute on Drug Abuse (NIDA) |
| Chemical compound, drug | BOL-148 | Other | NIDA; 7315–010 | National Institute on Drug Abuse (NIDA) |
| Chemical compound, drug | PZQ - praziquantel | Company | Sigma Aldrich: P4668-1G | |
| Chemical compound, drug | ergotamine | Company | Sigma Aldrich: 1241506–150 MG | |
| Chemical compound, drug | dihydroergotamine | Company | Sigma Aldrich: D1952000 | |
| Chemical compound, drug | bromocriptine | Company | Sigma Aldrich: B2134-100MG | |
| Chemical compound, drug | IBMX | Company | Sigma Aldrich: I5879-100MG | |
| Chemical compound, drug | LAMPA | Company | Cerilliant: L-004–1 ML | |
| Chemical compound, drug | Lisuride | Company | Tocris: 4052 | |
| Chemical compound, drug | dihydroergotoxine | Company | Tocris: 0474 | |
| Chemical compound, drug | methylergometrine | Company | Tocris: 0549 | |
| Chemical compound, drug | forskolin | Company | Cell Signaling Technology: 3828 | |
| Software, algorithm | HISAT2 | PMID: 25751142 | ||
| Software, algorithm | edgeR | PMID: 19910308 | ||
| Software, algorithm | Ingenuity Pathway Analysis | Company | QIAGEN Bioinformatics: IPA | |
| Software, algorithm | DAVID Bioinformatics Resources 6.8 | Other | Laboratory of Human Retrovirology and Immunoinformatics (LHRI) |
Additional files
-
Supplementary file 1
Natural product library screening data.
(Sheet 1) Vendor, chemical identification (SMILES) and primary screening data for all natural product compounds screened against Sm.5HTRL. (Sheet 2) Counter screening data for agonist primary hits in cells expressing the 22F cAMP GloSensor and lacking Sm.5HTRL. (Sheet 3) Counter screening data for antagonist primary hits in cells expressing the 22F cAMP GloSensor and lacking Sm.5HTRL. NPS = NCI Natural Products Set IV, NPL = TimTec Natural Product Library, NDL = TimTec Natural Derivatives Library.
- https://doi.org/10.7554/eLife.35755.017
-
Supplementary file 2
RNA-Seq data of drug treated and control infected mouse livers.
Read counts for uninfected control, infected control, infected ergotamine-treated and infected praziquantel treated liver samples.
- https://doi.org/10.7554/eLife.35755.018
-
Supplementary file 3
RNA-Seq data of drug treated and control infected mouse spleens.
Read counts for uninfected control, infected control, infected ergotamine-treated and infected praziquantel treated spleen samples.
- https://doi.org/10.7554/eLife.35755.019
-
Supplementary file 4
Signaling pathways enriched in drug treated livers relative to control infections.
Ingenuity Pathway Analysis (IPA) of transcripts differentially expressed in livers of PZQ (Sheet 1) and ergotamine (Sheet 2) treated mice relative to control infections. IPA generated activation Z-scores are shown if magnitude ≥2. KEGG pathway analysis of transcripts differentially expressed in livers of PZQ (Sheet 3) and ergotamine (Sheet 4) treated mice relative to control infections.
- https://doi.org/10.7554/eLife.35755.020
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35755.021





