Novel functions for integrin-associated proteins revealed by analysis of myofibril attachment in Drosophila
Figures
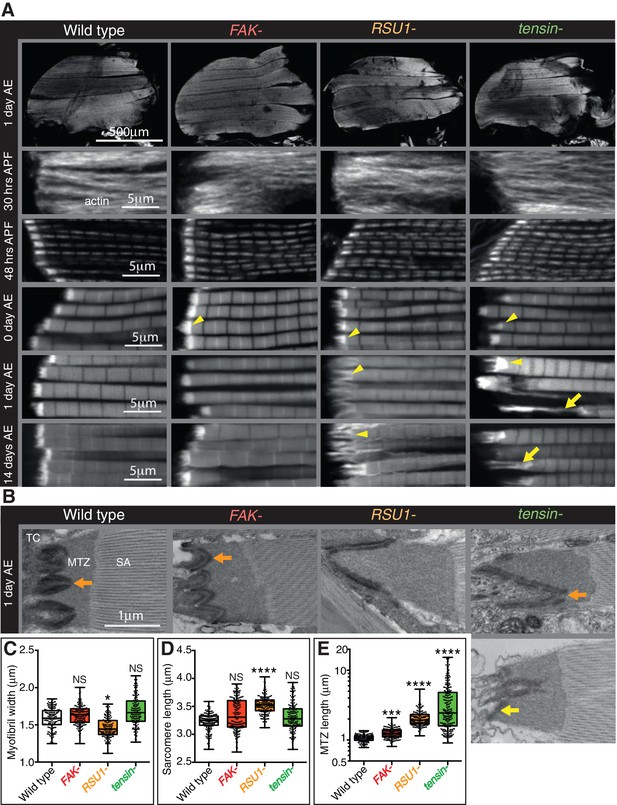
Indirect flight muscle (IFM) attachment sites reveal phenotypes for viable IAP mutants.
(A–E) Examination of IFMs in the absence of FAK, RSU1 and tensin. Samples were fixed at the stage indicated, stained for F-actin (white, using phalloidin in this and all subsequent figures), and imaged by confocal microscopy (in all cases except Figure 5). (A) Defects in MASs arise at eclosion of the adult from the pupal case (0 hr after eclosion (AE)), but are not observed during pupal stages (30 or 48 hr after pupae formation (APF)). Yellow arrowheads indicate myofibrils with elongated MTZs and yellow arrows indicate detaching myofibrils. Quantification of MTZ length is shown in E. (B) TEM images of MTZs at the indicated stage and genotype, showing MTZ, tendon cell (TC) and sarcomeric actin (SA). An electron dense region is observed close the plasma membrane, which is absent at the base of interdigitations in wildtype MTZs (11/12) (orange arrow) but is continuous in MTZs lacking FAK (9/10). In attached myofibrils, two electron dense lines are seen at the plasma membrane, whereas in detached myofibrils only one line is seen (yellow arrow). Note the disappearance of fine interdigitation in RSU1- and tensin -. (C–E) Quantitation of muscle parameters in adults 1 day AE: in these and all subsequent figures, for each genotype 10–30 myofibrils were measured from each of 10 individuals; all data points are shown, overlain with a boxplot showing median, interquartile range, minimum and maximum; statistical test used was Mann-Whitney U. *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001. (C) Myofibril width is smaller in the absence of RSU1 but not FAK or tensin. (D) Sarcomere length is longer in the absence of RSU1 but unchanged in the absence of FAK or tensin. (E) MTZ length is longer in all three mutants; full quantitation is in Figure 1—figure supplement 1. We used a log plot in this and subsequent graphs measuring MTZ length due to the great range in length.
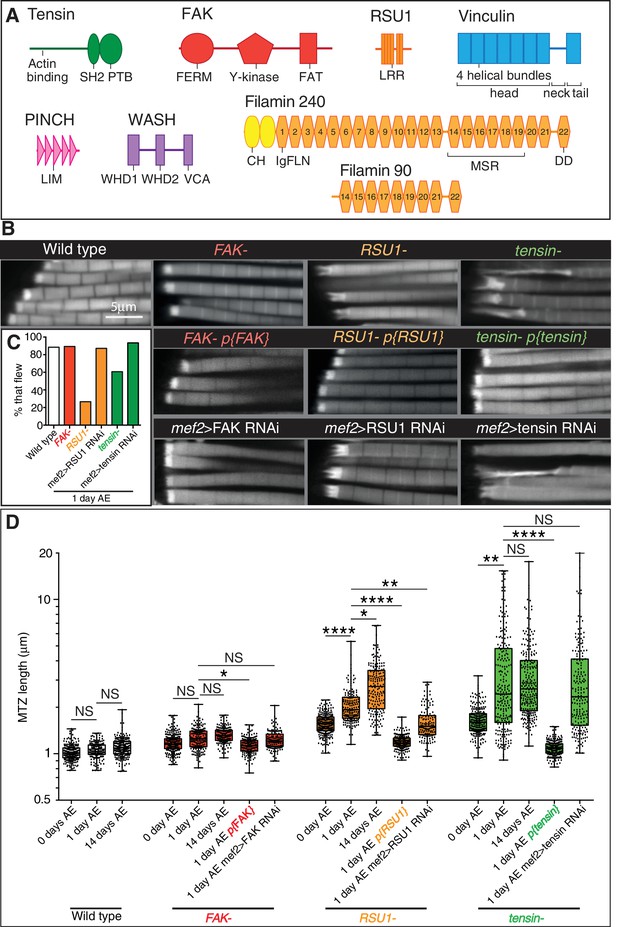
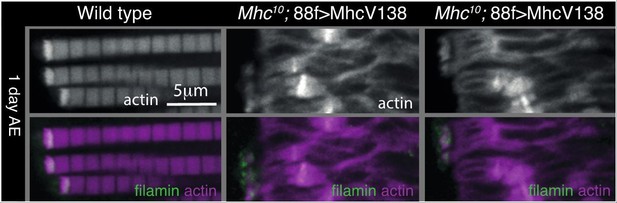
IAP mutant IFM defects are rescued by a copy of the IAP gene and phenocopied by RNAi in muscles.
(A) Domain structures for the proteins studied in this paper: tensin (green), FAK (red), RSU1 (orange), vinculin (blue), PINCH (pink), Wash (lilac), filamin (yellow). Drosophila tensin contains an actin binding region in its N-terminus and SH2 (Src Homology domain) and PTB (phospho-tyrosine binding) domains in its C-terminus but lacks the N-terminal PTP-like (phospho-tyrosine phosphatase) domain seen in vertebrate tensins. FAK contains a FERM (protein 4.1, ezrin, radixin and moesin homology) domain, tyrosine kinase domain (Y-kinase) and a focal adhesion targeting domain (FAT), which is thought to bind talin and paxillin. RSU1 is a leucine-rich repeat (LRR) containing protein. Vinculin is composed of three domains: a head domain, comprised of a series of four-helical bundles; a flexible proline-rich neck region; and a five-helical bundle tail domain. Vinculin head binds talin and α-actinin, the neck binds members of the CAP/vinexin family, and the tail binds actin, paxillin and the membrane lipid PIP2. PINCH contains 5 LIM domains, the most C-terminal of which binds RSU1. Wash contains two N-terminal Wash homology domains (WHD1 and 2) and a C-terminal VCA module which binds and activates the Arp2/3 complex. Drosophila filamin consists of an N-terminal actin binding domain comprising two CH (calponin homology) domains, and a series of immunoglobulin-like filamin repeats (IgFLN). The longest isoform (filamin240) contains 22 IgFLNs. IgFLN repeats 14–19 constitute the mechanosensor region (MSR) while the final IgFLN is the dimerisation domain (DD). The short isoform (filamin90) contains only the MSR and dimerization domain. (B) F-actin distribution in myofibrils of the genotypes indicated. Addition of genomic rescue constructs for each IAP rescues the phenotype. Driving RNAi in the muscles phenocopies the mutant phenotype. (C) Muscle defects in IAP mutants do not result in reduced flying ability. 30 flies per genotype were tested and percentage that fly is shown. (D) Quantitation of MTZ length as in Figure 1C.
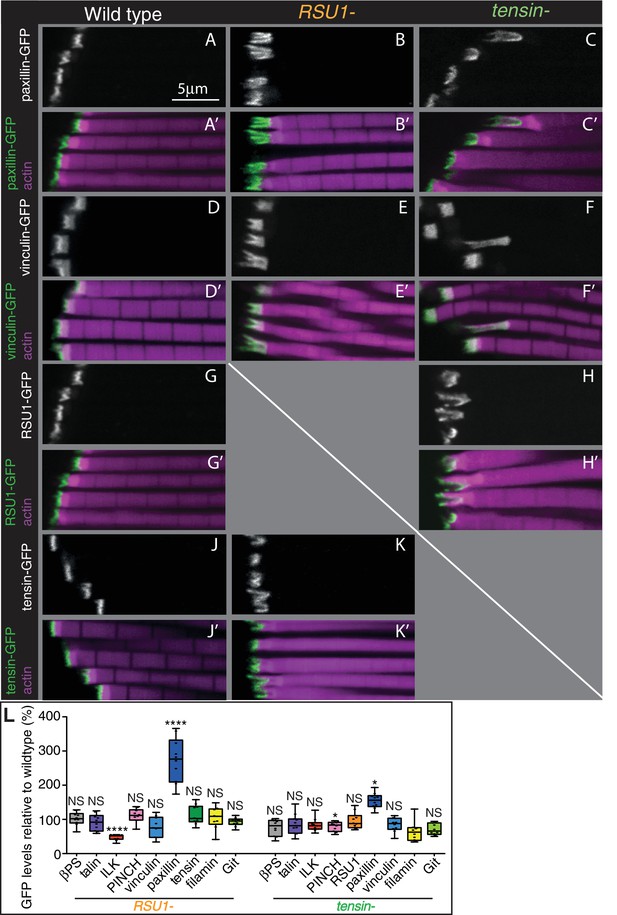
Other IAPs are still recruited in IFMs lacking RSU1 or tensin.
(A–K’) Confocal images showing the distribution of indicated GFP-tagged IAPs (white, green), each expressed from their own promoter, in the genotypes indicated at the top each panel. F-actin is magenta. The IFMs are from flies 1 day AE in this and all subsequent figures, unless indicated otherwise. (L) Quantification of IAP-GFP levels relative to levels in wild type IFMs at 1 day AE. For each genotype 10–30 myofibrils were measured from each of 10 individuals and an average from each individual was calculated; all data points are shown, overlain with a boxplot showing median, interquartile range, minimum and maximum; statistical test used was Mann-Whitney U. *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001 .
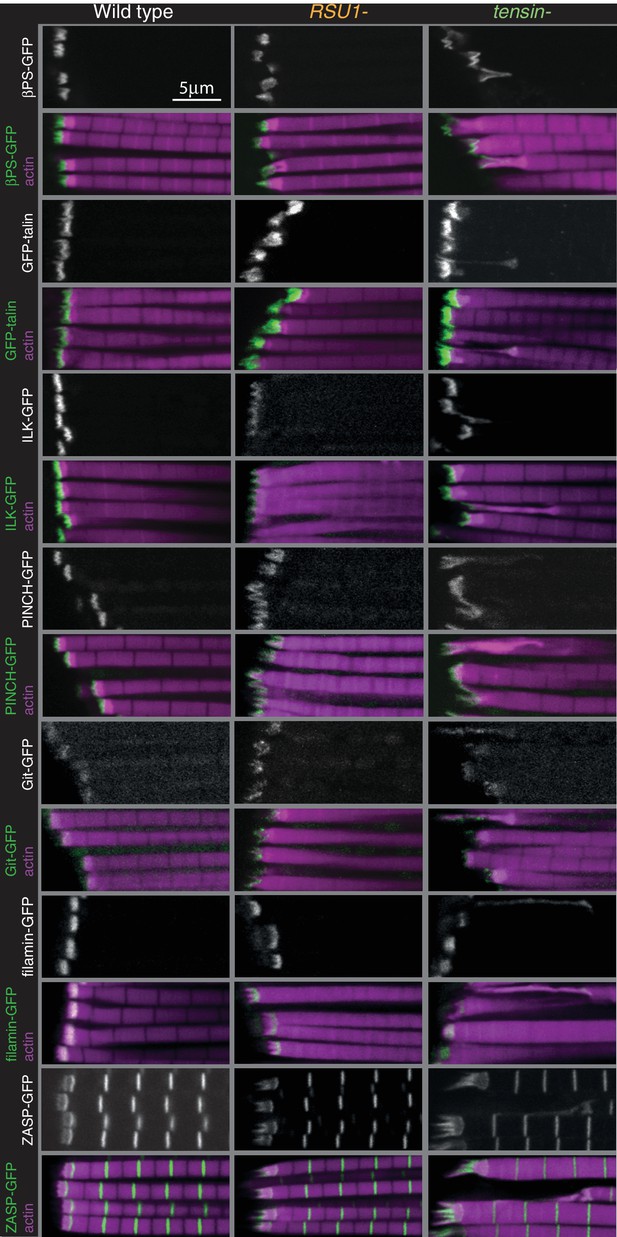
Distribution of additional IAPs in IFMs lacking RSU1 or tensin.
F-actin (magenta) and GFP-tagged IAPs (white, green), each expressed from their own promoter, in IFMs of genotypes indicated.
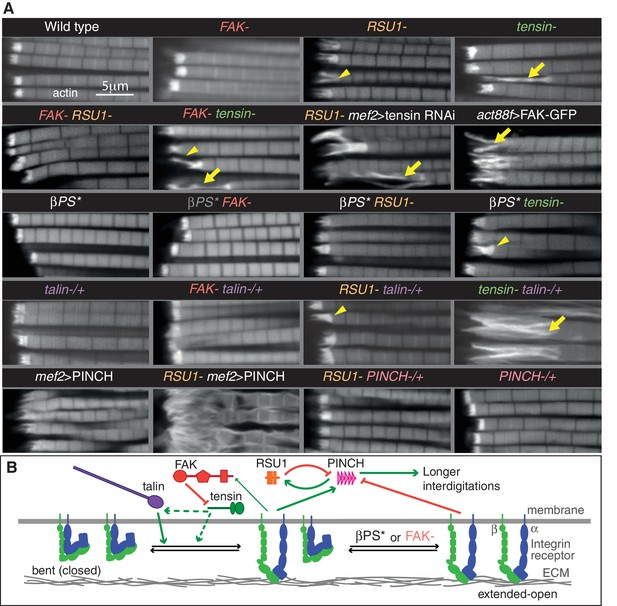
Epistasis reveals inhibitory action and compensation between IAPs.
(A) F-actin in myofibrils of the indicated genotype. βPS* is a missense mutant that makes βPS more active (mys[b27]). Yellow arrowheads indicate myofibrils with elongated MTZs and yellow arrows detaching myofibrils. Quantitation of MTZ length is in Figure 3—figure supplement 1. (B) Model of IAP function. Integrin heterodimers (green and blue) exist in an inactive bent conformation and an open upright conformation that binds ECM. Talin (purple) and tensin (green) both promote integrin activation, and FAK (red) inhibits activation by tensin. Active integrin performs adhesive functions through IAPs, including FAK and RSU1 (orange). RSU1 promotes adhesion by inhibiting excessive PINCH (pink) activity. Constitutively active integrin (βPS*) or loss of FAK, increases the amount of active integrins and stimulates the function of other IAPs that compensate for the loss of RSU1 or FAK.
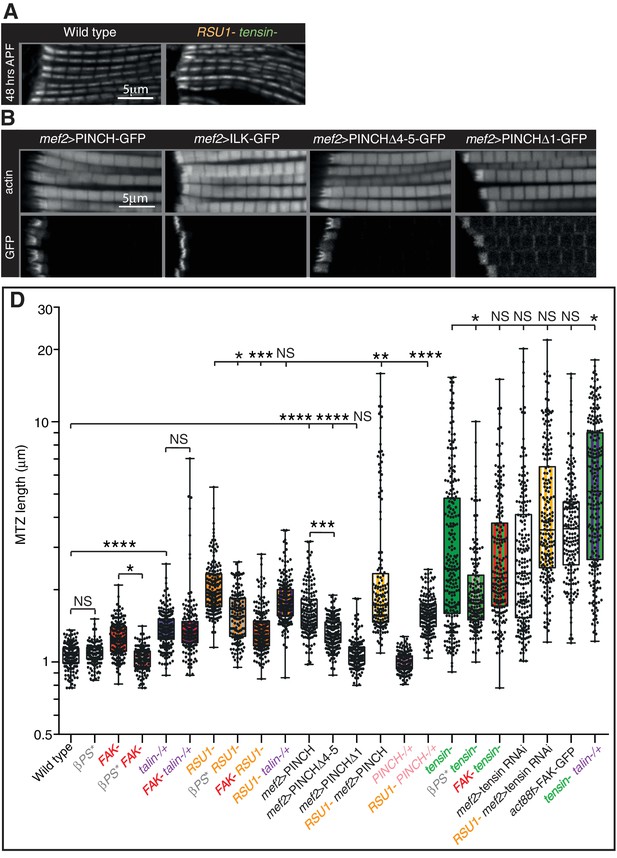
Defects are not enhanced in flies lacking both tensin and RSU1.
(A) IFM myofibrils in 48 hr APF pupae lacking both RSU1 and tensin appear normal despite the lethality during pupal stages. (B) IFMs overexpressing PINCHΔ4–5, but not PINCHΔ1 or ILK, phenocopy the overexpression of full length PINCH. Overexpressed PINCH and PINCHΔ4–5 localise are localised throughout the MTZ but enriched at the interdigitations. Conversely, ILK is only seen in interdigitations and PINCHΔ1 is not enriched at the interdigitations, suggesting that ILK is required for robust localisation of PINCH to the interdigitations. (C) Quantitation of the MTZ length of single and double mutants at 1 day AE shown in Figure 3 and Figure 3—figure supplement 1B.
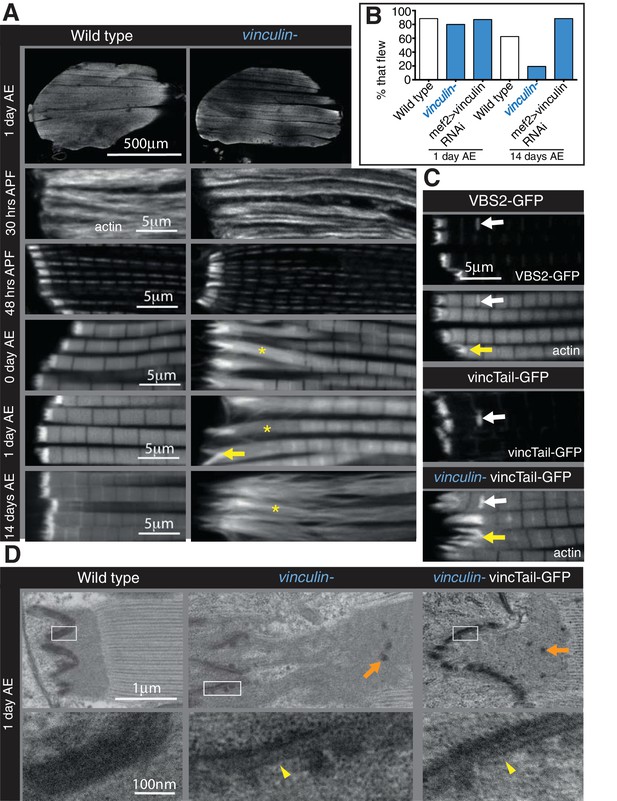
Integrin regulates actin structures at a distance, through vinculin.
(A) F-actin distribution (white) in wild type and vinculin- IFMs at the stages indicated. Yellow asterisks and yellow arrow indicate regions of disorganized actin. (B) Muscle defects do not result in reduced flying ability in the vinculin mutant. 30 flies per genotype were tested and percentage that fly is shown. (C) F-actin distribution (white) in IFMs of the genotype indicated. Vinculin tail and VBS2 were expressed with mef2-Gal4. White arrows indicate zebra bodies (abnormally large Z-lines). Yellow indicate detaching myofibrils. (D) TEM images of MTZs at the indicated stage and genotype. Orange arrows indicate electron dense aggregates in the MTZ. Yellow arrowheads indicate loss of electron dense material from the muscle, but not tendon cell, membrane.
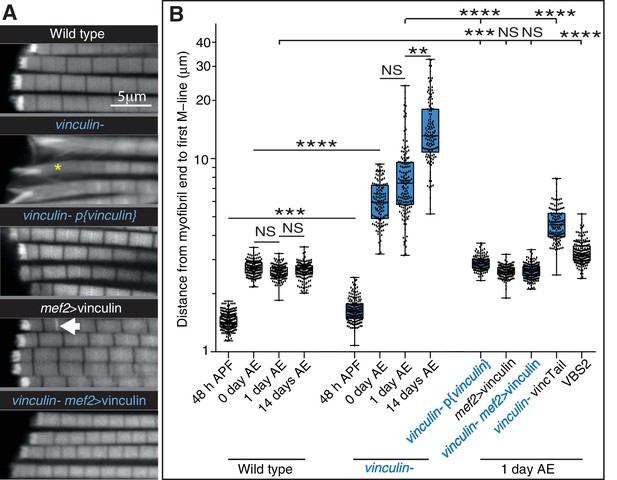
Defects seen with loss of vinculin are rescued by adding back wild type vinculin.
(A) F-actin distribution in IFMs of the genotypes indicated. The vinculin mutant phenotype is rescued by vinculin expressed either from its own promoter, p{vinculin}, or overexpressed with mef2-Gal4, mef2 >vinculin. Overexpression of vinculin alone induces some zebra bodies (white arrow). (B) Quantitation of the distance from myofibril end to first M-line. Note log scale.
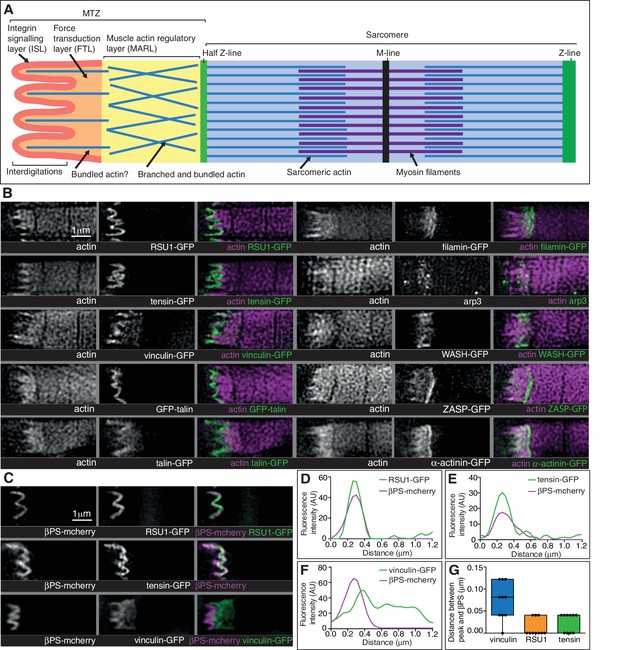
Super-resolution microscopy resolves distinct zones in the MTZ.
(A) Schematic of the four zones within the MTZ: (1) an integrin signalling layer (ISL, red) at the membrane; and then zones containing different actin structures-(2) a force transduction layer (FTL, orange); (3) a muscle actin regulatory layer (MARL, yellow); and (4) the first (half) Z-line (green) followed by the first sarcomere (blue). (B) SIM images of wild type myofibrils expressing IAPs tagged with GFP as indicated, or using antibody staining for arp3 (white and green), and stained for F-actin (white and magenta). Note the four zones: (1) the ISL containing tight membrane localization of RSU1, tensin, and GFP-talin tagged at the N-terminus and very little F-actin; (2) both ISL and FTL for vinculin and talin tagged toward the C-terminus; (3) the MARL containing filamin, vinculin, arp3 and WASH; (4) the first Z-line containing high levels of ZASP and α-actinin. Note that the web-like appearance of actin in the SIM images is an artefact of the SIM technique. See also Figure 5—figure supplement 1. (C–F) RSU1 and tensin overlap with integrin, in the ISL, whereas vinculin is separate, within the FTL. (C) SIM images of wild type myofibrils expressing pairs of proteins as indicated. (D–F) Representative line graphs, from images as in B, showing fluorescence intensity over distance. (G) Quantitation of the distance between the peaks of fluorescent intensity of βPS-mcherry versus RSU1-GFP, tensin-GFP or vinculin-GFP, combined from measurements of 10 myofibrils each. Boxplots show median, interquartile range, minimum and maximum.
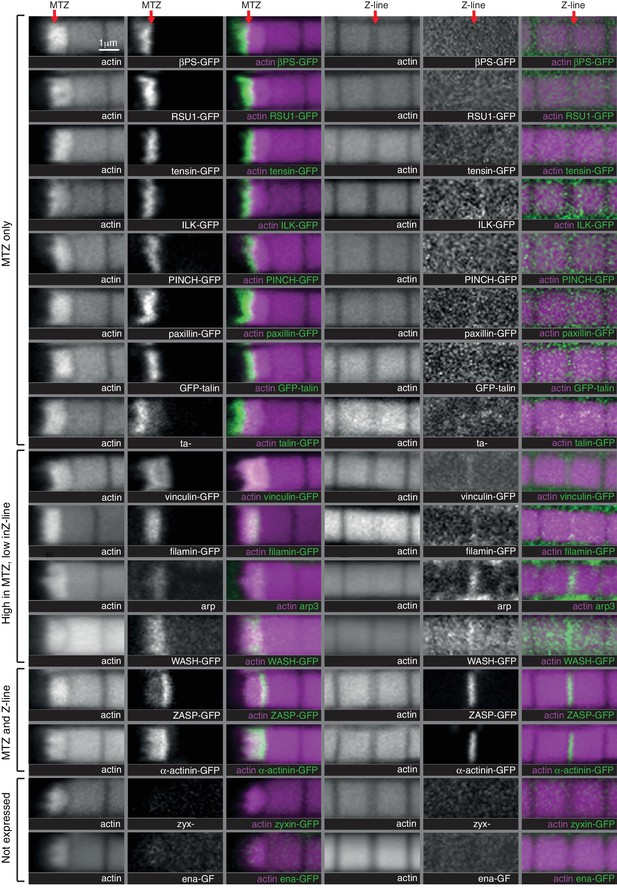
Comparison of IAP distribution at the MTZ versus Z-lines.
Distribution of F-actin (white, magenta) and GFP tagged proteins, or antibody staining for arp3, (white, green) in wild type IFMs, showing both the MTZ (left) and a representative Z-line in the middle of the myofibril (right). Note that levels at Z-lines are at much lower levels at Z-lines compare with the MTZ, and therefore images of Z-line distribution were taken at ~10X gain compared to the MTZ images, except ZASP and α-actinin, which were taken at the same settings. The proteins are enriched in different zones, indicated on the left.
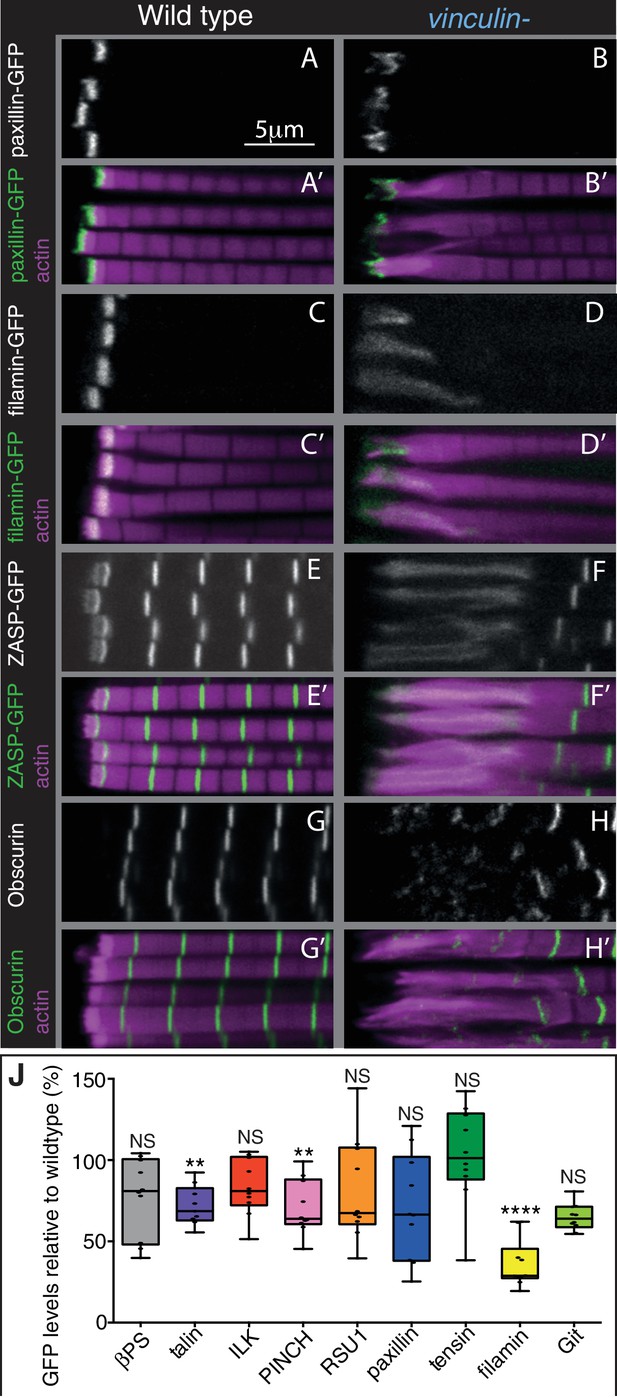
The absence of vinculin causes expansion of the MARL into the sarcomeres.
(A–H’) Distribution of F-actin (magenta) and the protein indicated (white, green) in wild type and vinculin- IFMs. MARL expansion is seen by expanded filamin-GFP and ZASP-GFP, and the underlying sarcomeres are visible with the M-line protein Obscurin/Unc-89 (antibody staining), whereas the ISL seems unaffected, as marked with paxillin-GFP. (J) Quantification of IAP-GFP levels relative to levels in wild type IFMs at 1 day AE. For each genotype 10–30 myofibrils were measured from each of 10 individuals and an average from each individual was calculated; all data points are shown, overlain with a boxplot showing median, interquartile range, minimum and maximum; statistical test used was Mann-Whitney U. *p≤0.05; **p≤0.01; ***p≤0.001; ****p≤0.0001.
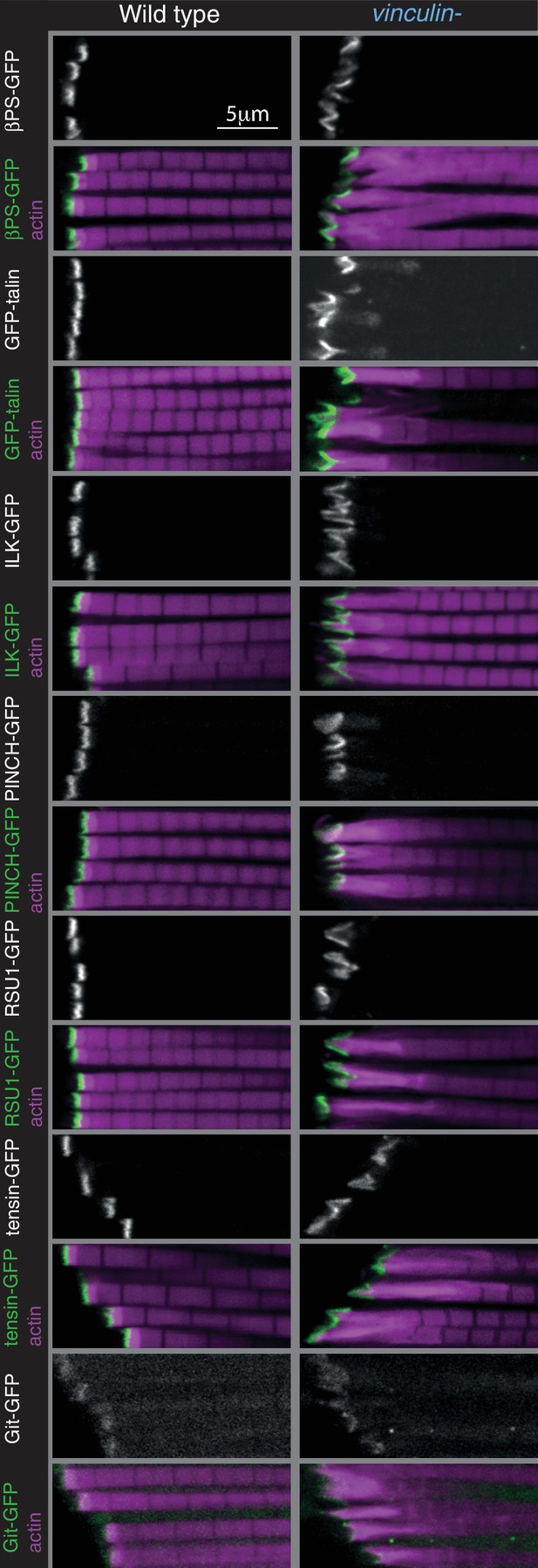
Distribution of additional IAPs in IFMs lacking vinculin.
Distribution of F-actin (white, magenta) and GFP tagged IAPs (white, green) in IFMs. GFP-talin moves into the expanded MARL in the absence of vinculin; otherwise IAP distribution appears normal.
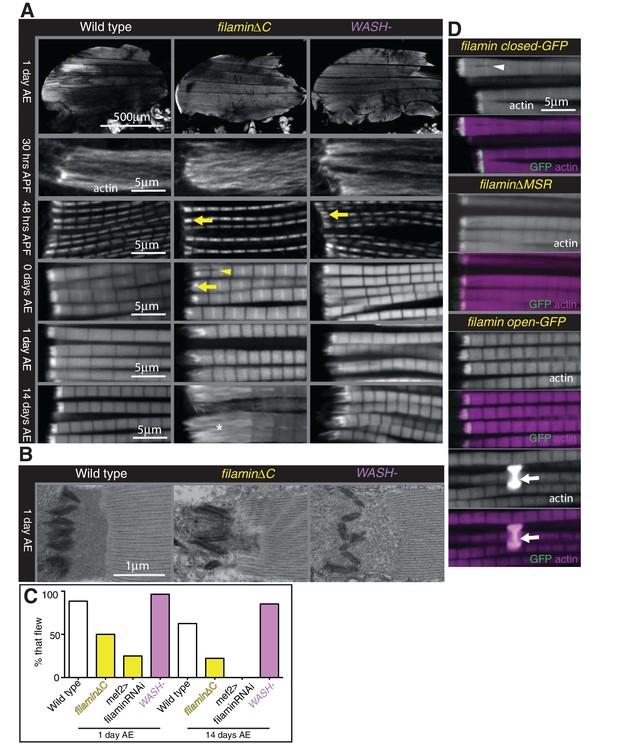
Filamin and WASH contribute to MARL formation.
(A) Loss of filamin or WASH results in a smaller MARL at 48 hr APF and 0 days AE, followed by an expansion of the MARL at later stages. F-actin distribution (white) of the genotypes and stages as indicated. Yellow arrows indicate small MTZs, asterisks indicate expanded and disorganised MARL, and yellow arrowheads indicate split myofibrils observed in these mutants but not vinculin (Figure 4). (B) TEM images of MTZs at the indicated stage and genotype. (C) Muscle defects caused by loss of filamin, but not WASH, lead to reduced flying ability. 30 flies per genotype were tested and percentage that fly is shown. (D) Mechanical opening of filamin is required for MARL formation. F-actin and filamin-GFP distribution. Filamin that is easier to open makes a normal MTZ, and ectopic zebra bodies (white arrows). White arrowheadindicates split myofibrils.
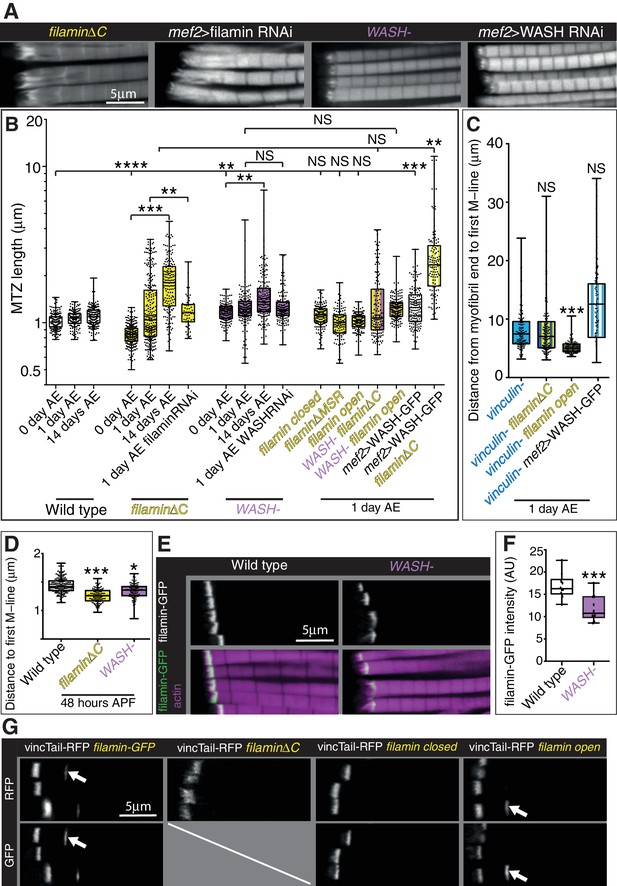
Filamin and WASH contribute to MARL formation.
(A) Distribution of F-actin in IFMs of the genotypes indicated. (B) Quantitation of MTZ length of genotypes shown in Figure 7. (C) Quantitation of MARL expansion for the genotypes shown in Figure 7C. Filamin open rescues loss of vinculin. (D) Quantitation of MARL reduction during pupal stages, 48 hr APF, of genotypes in Figure 7A. (E,F) Reduction of filamin (green) levels in the absence of WASH. (F) Quantitation of filamin-GFP levels at the MTZ. (G) Ability of vinculin tail to form zebra bodies requires the mechanical opening of filamin. F-actin (white, magenta) and vinculin tail-RFP (green) in IFMs of the genotype indicated at 1 day AE. White arrows indicate zebra bodies. If filamin is removed or stabilized in the closed conformation, zebra bodies are not observed.
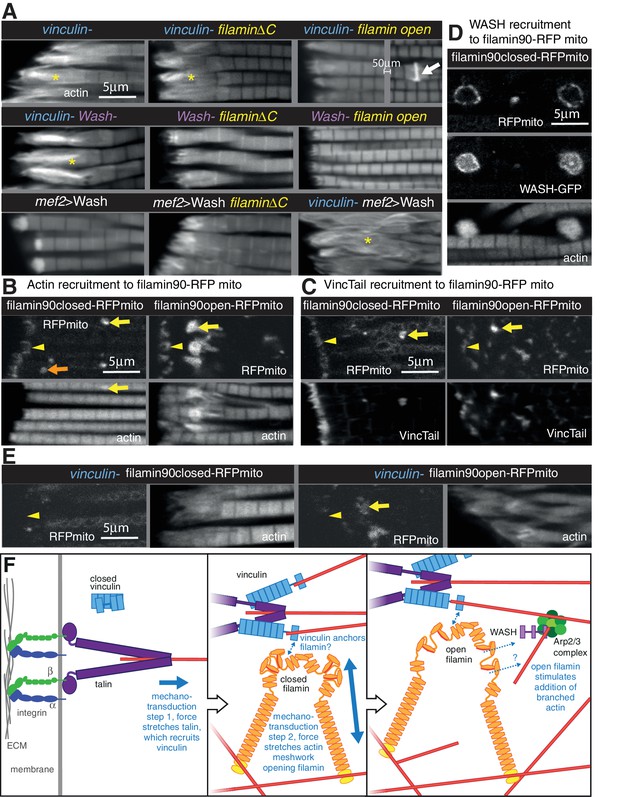
Vinculin, filamin and WASH work together to build the MARL.
(A) F-actin distribution of the genotypes indicated. Yellow asterisks indicate disorganised actin. Combining mutants does not enhance the phenotypes, filamin open rescues absence of vinculin but not WASH, and overexpression of WASH expands the MTZ and enhances loss of filamin. White arrow indicates zebra bodies. (B) Filamin90open recruits actin more robustly than filamin90closed. Yellow arrows indicate mitochondrial aggregates that rcruit actin, orange arrows indicate aggregates that do not recruit actin and yellow arrowheads indicate the MTZ. (C) Filamin90open and closed-RFPmito recruit vinculinTail. (D) Filamin90closed recruits WASH-GFP, leading to large spheres of F-actin. (E) Vinculin is required for recruitment of filamin90-RFPmito constructs to the MTZ. (F) Model of MARL formation by vinculin (blue), filamin (yellow) and WASH (lilac). Mechanical stretching of talin (purple) reveals cryptic binding sites for vinculin. Vinculin binding to talin opens up vinculin, revealing it’s actin binding tail. The tail anchors filamin to actin filaments causing the MSR of filamin to be stretched. Open filamin then stimulates addition of branched actin filaments through WASH and Arp2/3 (green) activity.
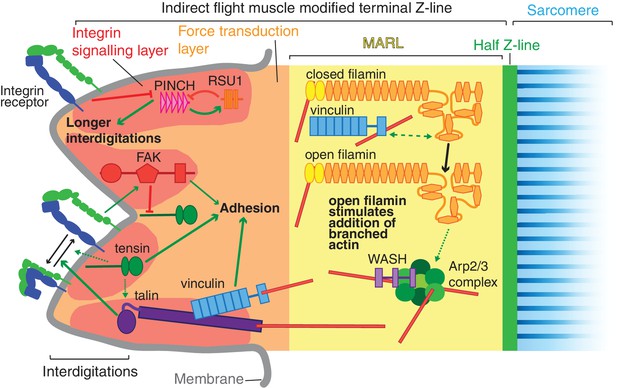
Model of IAP function in the IFM MTZ.
The MTZ is composed of 4 zones: (1) an integrin signalling layer (ISL, red) at the membrane; and then zones containing different actin structures-(2) a force transduction layer (FTL, orange); (3) a muscle actin regulatory layer (MARL, yellow); and (4) the first (half) Z-line (green) followed by the first sarcomere (blue). Within the ISL, RSU1 (orange), PINCH (pink), tensin (green) and FAK (red) regulate integrin adhesion. Tensin promotes integrin activation, possibly through talin, and this activity is inhibited by FAK. Excessive PINCH activity, which leads to longer interdigitations than wildtype, is inhibited by RSU1. Within the FTL, stretched talin recruits and opens up vinculin, revealing its actin binding tail. In the MARL, vinculin tail anchors filamin to actin, leading to stretching of filamin and opening up of its mechanosensor region. Open filamin promotes addition of branched actin, possible through activation of WASH and the Arp2/3 complex.
Tables
| Reagent type (species) or Resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | rabbit anti-obscurin | (Burkart et al., 2007) | N/A | |
| Antibody | rabbit anti-arp3 | (Stevenson et al., 2002) | N/A | |
| Antibody | rat anti-filamin C terminus | (Sokol and Cooley, 1999) | N/A | |
| Genetic reagent (Drosophila melanogaster) | FAK-: FakCG | (Grabbe et al., 2004) | N/A | |
| Genetic reagent (Drosophila melanogaster) | RSU1-: ics2 | This paper | N/A | Null allele for ics (RSU1) |
| Genetic reagent (Drosophila melanogaster) | tensin-: by33c | (Torgler et al., 2004) | N/A | |
| Genetic reagent (Drosophila melanogaster) | vinculin-: ΔVinc | (Klapholz et al., 2015) | N/A | |
| Genetic reagent (Drosophila melanogaster) | filamin/cheerioΔC: chers24 | (Huelsmann et al., 2016) | N/A | |
| Genetic reagent (Drosophila melanogaster) | filamin/cheerio open: cheropen MSR | (Huelsmann et al., 2016) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | filamin/cheerio closed: cherclosed MSR | (Huelsmann et al., 2016) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | filamin/cheerio ΔMSR: cherΔMSR | (Huelsmann et al., 2016) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | βPS*: mysb27 | (Kendall et al., 2011) | N/A | |
| Genetic reagent (Drosophila melanogaster) | talin-: rhea62 | (Klapholz et al., 2015) | N/A | |
| Genetic reagent (Drosophila melanogaster) | WASH-: Wash185* | (Nagel et al., 2017) | N/A | |
| Genetic reagent (Drosophila melanogaster) | βPS-GFP: mys-GFP | (Klapholz et al., 2015) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | GFP-talin | (Klapholz et al., 2015) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | talin-GFP | (Klapholz et al., 2015) | N/A | Fully functional gene trap |
| Genetic reagent (Drosophila melanogaster) | Paxillin-GFP: | (Bataillé et al., 2010) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | Git-GFP: | (Bulgakova et al., 2017) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | ILK-GFP: | (Zervas et al., 2001) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | PINCH-GFP: | (Kadrmas et al., 2004) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | RSU1-GFP: | This paper | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | tensin-GFP: | (Torgler et al., 2004) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | vinculin-GFP: | (Klapholz et al., 2015) | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | ZASP-GFP: | (Jani and Schöck, 2007) | N/A | Fully functional gene trap |
| Genetic reagent (Drosophila melanogaster) | α-actinin-GFP: | (Buszczak et al., 2007) | N/A | Fully functional gene trap |
| Genetic reagent (Drosophila melanogaster) | filamin/cheerio-GFP: cher-GFP | Huelsmann et al., 2016) | N/A | Viable line made by homologous recombination |
| Genetic reagent (Drosophila melanogaster) | UAS-WASH-GFP | (Nagel et al., 2017) | N/A | |
| Genetic reagent (Drosophila melanogaster) | zyxin-GFP: | This paper | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | ena-GFP: | Kindly provided by Yoshiko Inoue, Sven Huelsmann and Jenny Gallop | N/A | Genomic rescue construct |
| Genetic reagent (Drosophila melanogaster) | UAS-vinculin-RFP: | (Maartens et al., 2016) | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-vinculinTail-RFP: | (Maartens et al., 2016) | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-VBS2-GFP: | (Maartens et al., 2016) | N/A | |
| Genetic reagent (Drosophila melanogaster) | mef2-Gal4: P{Gal4-Mef2.R}3 | Bloomington Drosophila stock center | BDSC:50742 | |
| Genetic reagent (Drosophila melanogaster) | act88f-Gal4: P{Gal4-act88f} | (Bryantsev et al., 2012) | N/A | |
| Genetic reagent (Drosophila melanogaster) | y1 P(nos-cas9, w+) M (3xP3-RFP.attP)ZH-2A w* | Bloomington Drosophila stock center | BDSC:54591 | |
| Genetic reagent (Drosophila melanogaster) | FAK-RNAi: | Bloomington Drosophila stock center | BDSC:44075 | |
| Genetic reagent (Drosophila melanogaster) | RSU1-RNAi: | Vienna Drosophila Resource centre | VDRC:42188 | |
| Genetic reagent (Drosophila melanogaster) | tensin-RNAi: | Vienna Drosophila Resource centre | VDRC:22823 | |
| Genetic reagent (Drosophila melanogaster) | filamin-RNAi: | Vienna Drosophila Resource centre | VDRC:107451 | |
| Genetic reagent (Drosophila melanogaster) | WASH-RNAi: | Vienna Drosophila Resource centre | VDRC:24642 | |
| Genetic reagent (Drosophila melanogaster) | UAS-ILK-GFP: | (Zervas et al., 2011) | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-PINCHΔ1-GFP: | (Zervas et al., 2011) | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-PINCHΔ4–5-GFP | This paper | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-filamin90closed- RFPmito: | This paper | N/A | |
| Genetic reagent (Drosophila melanogaster) | UAS-filamin90open- RFPmito: | This paper | N/A | |
| Oligonucleotide | 5’-GTCGCTTCAAGAA CCCCATGTATG-3’ | This paper | N/A | Guide RNA for mys-mcherry |
| Oligonucleotide | 5’-AAACCATACATGG GGTTCTTGAAG-3’ | This paper | N/A | Guide RNA for mys-mcherry |
| Oligonucleotide | 5’-ACGCGTGGGAAT AGCAAACGCCACA-3’ | This paper | N/A | Primer to amplify 5’UTR of zyxin |
| Oligonucleotide | 5’-TGTGTACTTGCG CATTCACA-3’ | This paper | N/A | Primer to amplify 5’UTR of zyxin |
| Oligonucleotide | 5’-GACGTCAGAACA TTCGAGCTCATCGAT GAGTAAAGGA-3’ | This paper | N/A | Primer to introduce Aat2 site to GFP |
| Oligonucleotide | 5’ TGCGCAATAAATAAAAT GAGCACTCAATTTATTTGT ATAGTTCATCCATGC-3’ | This paper | N/A | Primer to add Fsp1 site to GFP |
| Recombinant DNA reagent | pBluescript II KS | (addgene) | X52327.1 | |
| Recombinant DNA reagent | pBluescript II KS_mys- mcherry | This paper | N/A | Plasmid for generation of mys-mcherry flies |
| Recombinant DNA reagent | pCFD3 | (Port et al., 2014) | N/A | |
| Recombinant DNA reagent | BACR13D24 | (Berkeley Drosophila genome project) | AC010838 | |
| Recombinant DNA reagent | TOPO TA | (Thermo Fisher Scientific) | 451641 | |
| Recombinant DNA reagent | pWhiteAttPRabbit zyxin-GFP | This paper | N/A | Plasmid for generation of zyxin-GFP flies |
| Recombinant DNA reagent | pWhiteAttPRabbit RSU1-GFP | This paper | N/A | Plasmid for generation of RSU1-GFP flies |
| Recombinant DNA reagent | pUASP-attB | Drosophila Genomics Resource center | 1358 | |
| Recombinant DNA reagent | MrRFPmito | (Maartens et al., 2016) | N/A | |
| Recombinant DNA reagent | pUASP-filamin90closed -RFPmito | This paper | N/A | Plasmid for generation of filamin90closed-RFPmito flies |
| Recombinant DNA reagent | pUASP-filamin90open -RFPmito | This paper | N/A | Plasmid for generation of filamin90open-RFPmito flies |
| Software | FIJI (ImageJ v1.5) | NIH | RRID:SCR_002285 | |
| Software | softWoRx 6.0 | Applied Precision | ||
| Software | Prism | GraphPad | RRID:SCR_002798 | |
| Software | Adobe Photoshop | Adobe | RRID:SCR_014199 | |
| Software | Microsoft Excel | Microsoft | RRID:SCR_016137 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35783.019





