Structural insights into the architecture and membrane interactions of the conserved COMMD proteins
Figures
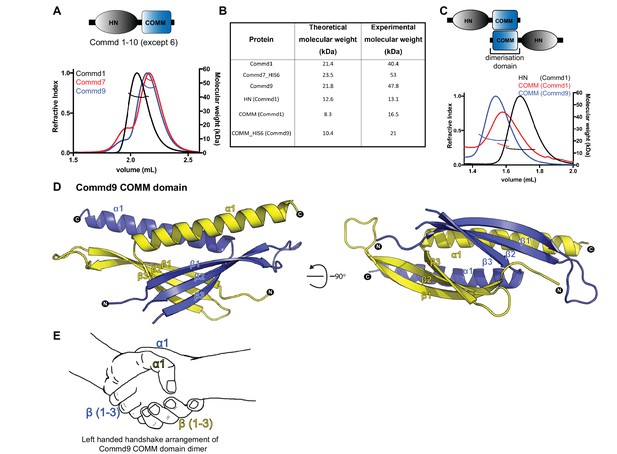
COMMD proteins dimerize through the C-terminal COMM domain.
(A) Cartoon representation of COMMD proteins, and the MALLS profile of Commd1 (black), Commd7 (red) and Commd9 (blue) showing Commd proteins are dimers in solution (B) Comparison of the theoretical molecular weight of the COMMD proteins and the experimentally measured molecular weight. (C) MALLS analyses of the COMM and HN domain shows that the protein dimerisation occurs through the C-terminal COMM domain of the COMMD proteins as represented schematically. (D) Ribbon representation of the dimeric Commd9 COMM domain (residues 115–198) chain A (yellow) and chain B (blue). (E) The structure of the COMM domain dimer is analogous in orientation to a left-handed handshake.
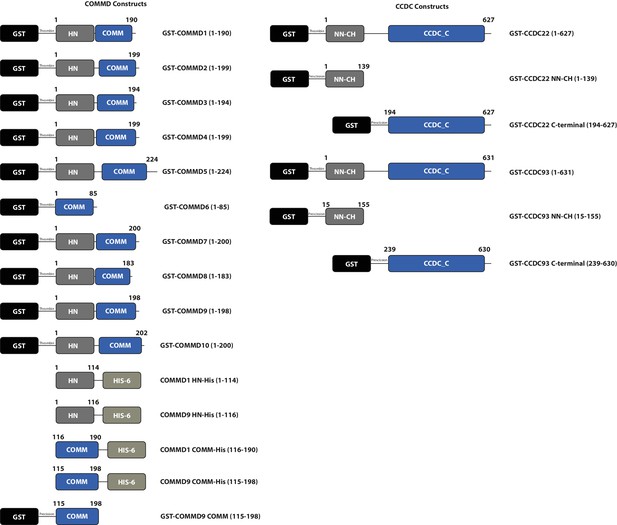
Schematic representations of bacterial expression constructs used in this study.
https://doi.org/10.7554/eLife.35898.003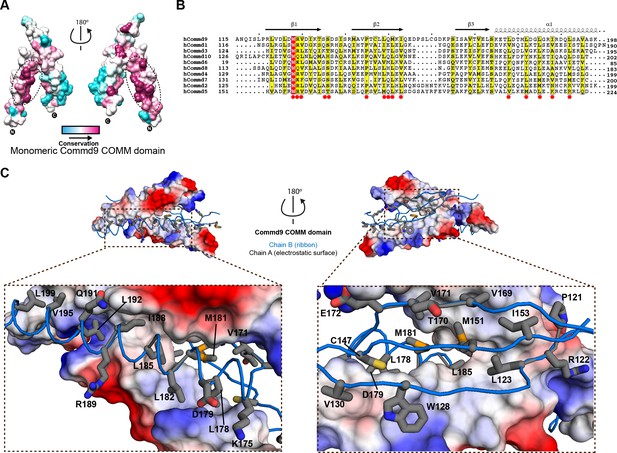
Structural mechanism underpinning the COMMD dimerization.
(A) A surface map of the conserved and variable residues of the Commd9 COMM domain showing the hydrophobic core is highly conserved while the surface residues are more variable, confirming the importance of dimerization for COMMD stability. These calculations are made using the Consurf server (Ashkenazy et al., 2010) on chain A. (B) A combined sequence alignment and secondary structure comparison of COMM domains of all the human COMMD proteins highlights that the C-terminal COMM domain is highly conserved across the COMMD family of proteins. Residues marked by asterisk depict conservation of amino acids that decorate the dimerization interface. Alignments were made with ESPript 2.2 (http://espript.ibcp.fr/ESPript/ESPript/) (Gouet et al., 2003) (C) Representation of dimer interface of Commd9 COMM domain highlighting key residues (chain A, worm in blue) making main chain-main chain, stacking, salt bridge interactions with the electrostatic surface (chain B).
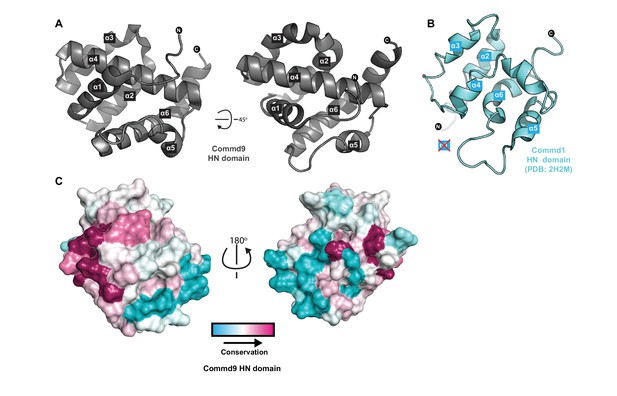
Structure of the N-terminal HN domain of Commd9.
(A) Cartoon representation of Commd9 HN domain (1–116) crystal structure crystallized at pH 4.9 showing a globular structure. (B) NMR structure of the Commd1 HN domain (PDB ID 2H2M) (Sommerhalter et al., 2007) shown in cartoon diagram in the same orientation as Commd9 in Figure 3A left panel. The Commd1 HN domain lacks the first helix (α1) and appears to have a kinked helix α4, but overall shares the same topology. (C) Sequence conservation of Commd9 HN domain mapped on the structure using CONSURF.
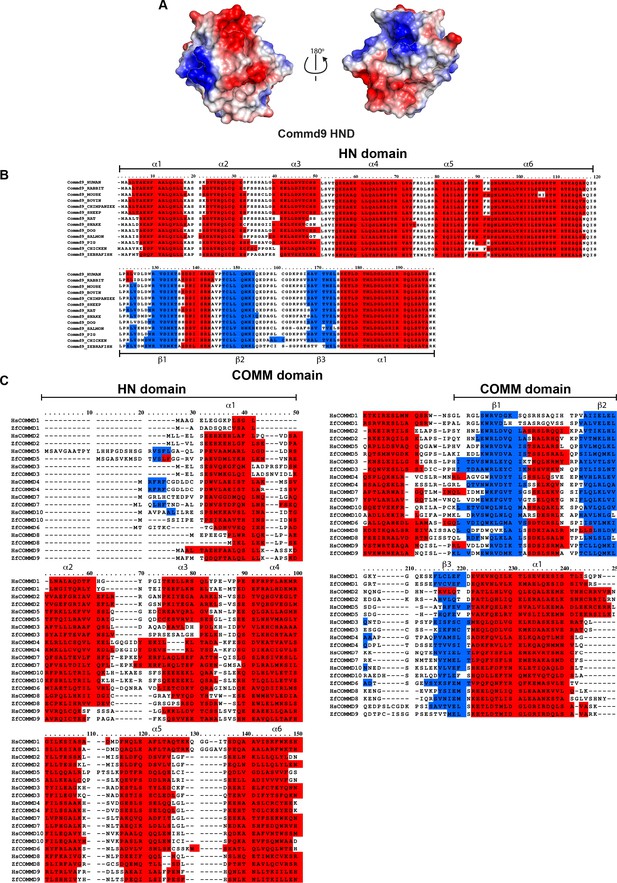
Sequence alignments of HN domain of all the human and zebrafish COMMD proteins.
(A) Electrostatic potential surface of Commd9 HN domain. (B) Species wide multiple sequence alignment of Commd9 showing conserved HN domain and COMM structural modules. (C) Multiple sequence alignment of human and zebrafish COMMD proteins.
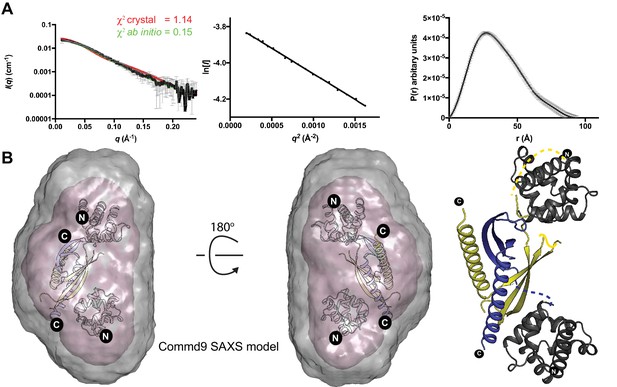
The solution structure of Commd9 determined by SAXS.
(A) (Left) Experimental scattering profile Commd9 in black overlaid with the theoretical scattering curve calculated from the Commd9 rigid body model (red) and the ab initio model determined by GASBOR (green) using CRYSOL. (Middle) The Guinier plot for the experimental data at the low-angle region (qmax × Rg < 1.3). (Right) P(r) functions derived from the SAXS data. (B) (Left) Averaged (grey) and filtered (coral) molecular envelopes from GASBOR. The ab initio model of Commd9 was docked with the rigid body model of Commd9 using SUPCOMB. (Right) Model of Commd9 protein from the experimental SAXS shown in ribbon diagram with HN domains in grey and the COMM domain dimer in blue and yellow.
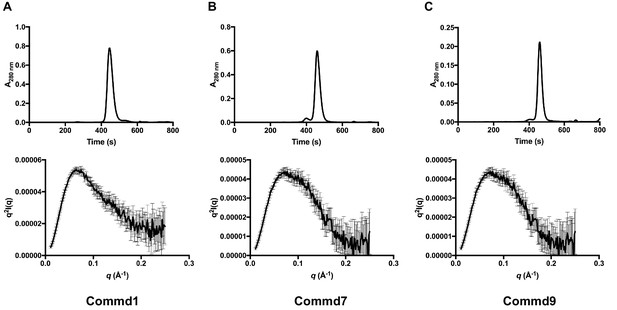
SEC-SAXS profiles of Commd1, Commd7 and Commd9.
(A) SEC-SAXS chromatogram (top) and Kratky plot (bottom) of Commd1, (B) Commd7 and, (C) Commd9.
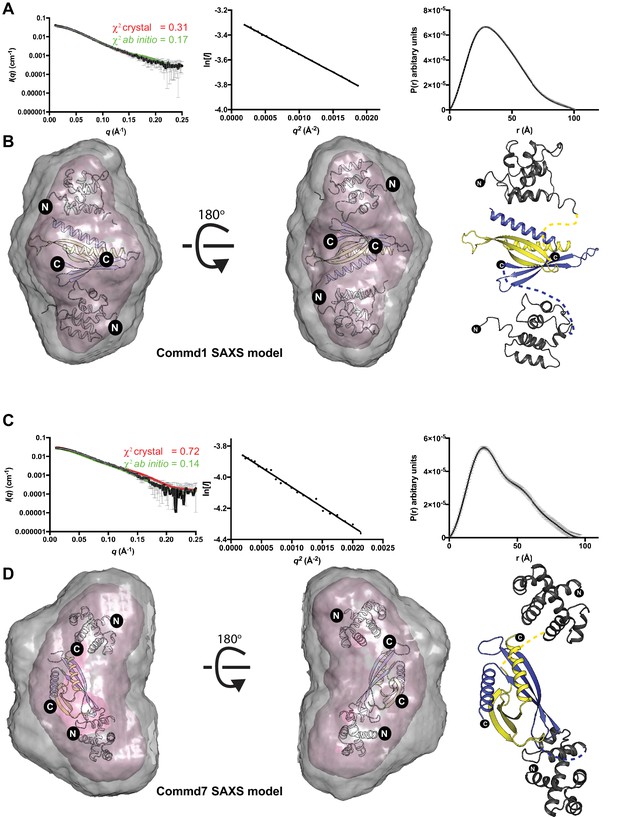
The solution structures of Commd1 and Commd7 determined by SAXS.
(A, C) (Left) Experimental scattering profiles of Commd1 and Commd7 in black overlaid with the theoretical scattering curves calculated from the Commd1 and Commd7 rigid body model (red) and the ab initio model determined by GASBOR (green) using CRYSOL. (Middle) The Guinier plot for the experimental data at the low-angle region (qmax × Rg < 1.3). (Right) P(r) functions derived from the SAXS data. (B, D) (Left) Averaged (grey) and filtered (coral) molecular envelopes from GASBOR. The ab initio models of Commd1 and Commd7 were docked with rigid body models of Commd1 and Commd7 respectively using SUPCOMB. (Right) Models of Commd1 and Commd7 generated from the experimental SAXS data shown in ribbon diagram with HN domains in grey and the COMM domain dimer in blue and yellow.
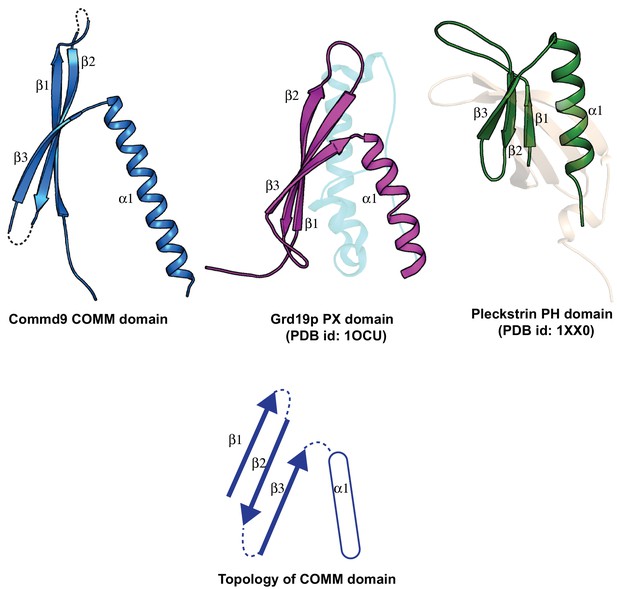
Structural comparison of the COMM domain of Commd9 with other similar modules.
https://doi.org/10.7554/eLife.35898.011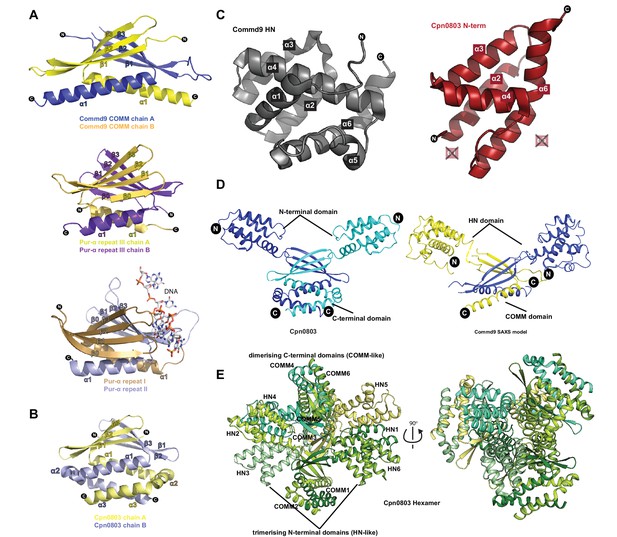
Structural similarity of COMMD proteins to Pur-α repeats and chlamydial proteins.
(A) Ribbon representation of Commd9 COMM domain homodimer compared to the structures of Pur-α repeats (PDB ID 5FGO, 5FGP) (Graebsch et al., 2009; Weber et al., 2016). Pur-α repeat III forms a homodimer similar to Commd9, but possesses an additional β-strand at the N-terminus. Repeats I and II from Pur-α form an intramolecular ‘heterodimer’ that is responsible for binding to DNA (shown in sticks). (B) The structure of the C-terminal domain of the C. pneumoniae Cpn0803 protein (PDB ID 3Q9D) (Stone et al., 2012) is shown in ribbon diagram. The C-terminal domain forms a homodimer similar to the Commd9 COMM domain, but possesses two additional C-terminal α-helices. C. trachomatis protein CT584 has an identical structure (PDB ID 4MLK) (Barta et al., 2013). (C) Side by side view of Commd9 HN domain, and the N-terminal domain of Cpn0803 (residues 8–98) showing the similarity in the overall secondary structure topology. The chlamydial proteins lack helices corresponding to Commd9 α1 and α5 (D) Comparison of the full-length Commd9 structure determined by SAXS and the Cpn0803 crystal structure, highlighting the similarity in their domains and their inter-domain orientations. (E) Ribbon representation of Cpn0803 showing the dimerization (COMM domain-like) module and the trimerisation (HN domain like) domain.
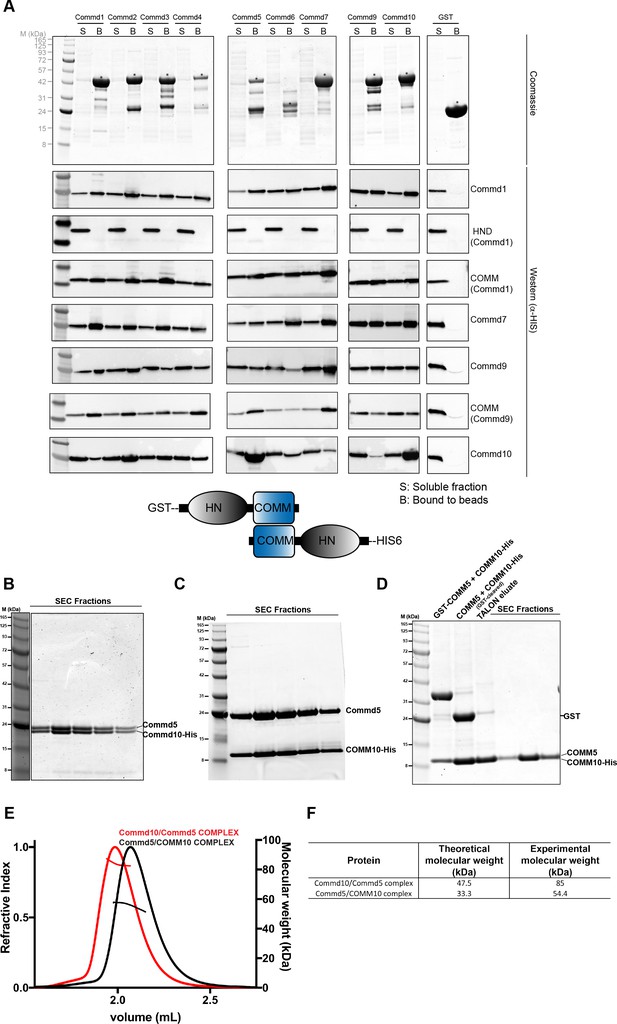
COMMD proteins form an array of homo and heterodimers via the C-terminal COMM domain.
(A) A representative Coomassie image showing the relative expression of the GST fusion COMMD proteins (top). Blots probed with anti-His6 antibody demonstrating the direct pair-wise interactions of the COMMD protein family (bottom). A schematic representation of COMMD-COMMD interaction as demonstrated by the pull-downs. (B, C, D) Representative Coomassie images showing the reconstitution of full length Commd5-Commd10 complex, full length Commd5-Comm domain of Commd10 complex and complex of COMM domains of Commd5 and Commd10 respectively. (E, F) MALLS analyses of full length Commd5-Commd10 complex and Commd5-COMM domain of Commd10 complex showing the complexes potentially form heterotetramers based on the calculated molecular weight of the complexes.
Raw files of SDS-PAGE gels and western blots of COMMD-COMMD and COMMD-CCDC interactions.
https://doi.org/10.7554/eLife.35898.015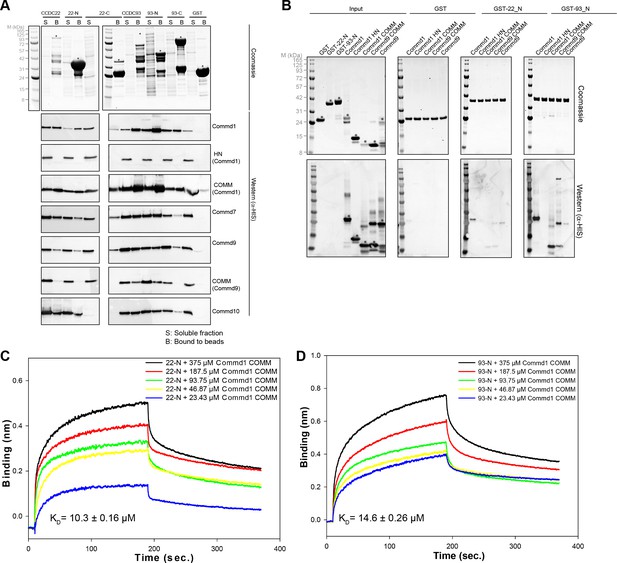
COMMD proteins bind directly to the N-terminal NN-CH domains of CCDC22 and CCDC93 via the COMM domain.
(A) A representative coomassie gel of GST fusion protein expression in a co-expression set-up (Top). Anti-His6 blots of the GST pull down demonstrating direct interactions between the COMMD family proteins and the Coiled-coil domain proteins 22 and 93 (Bottom). (B) GST pull down demonstrating the binding capacity of the N-terminal domain of CCDC22 and 93 to Commd1 and Commd9 as well as their respective COMM domains. (C and D) Binding of the Commd1 COMM domain to the N-terminal domains of CCDC22 (C) and CCDC93 (D) measured at varying concentrations using the BLiTz system. Binding kinetics we calculated using Sigma Plot (Systat Software Inc.).
COMMD proteins bind to NN-CH domain of CCDC93 via the conserved WRVD motif in the COMM domain.
(A) Cross-linking of Commd9, NN-CH domain of CCDC93 and the mixture of two with BS3 for 30 min at room temperature. Three distinguishable SDS-PAGE gel bands were excised in the complex mixture sample (indicated by the number 1, 2 and 3) for MS analysis. In parallel, a cross-linking reaction between Commd9 and lysozyme was performed under the same condition. (B) Cross-link map for Commd9 in complex with the NN-CH domain of CCDC93. Intermolecular and intramolecular cross-linked peptides are labelled as black and brown dot lines respectively. (C and D) Ribbon representation of the Commd9 HN (C) and (D) COMM domain structures with mapped intramolecular and intermolecular cross-linked lysine residues (in spheres). In the Commd9 COMM domain, K133 and K152 are part of a contiguous surface that includes the side-chains of the conserved residues 128WRVD131 (indicated with sticks).
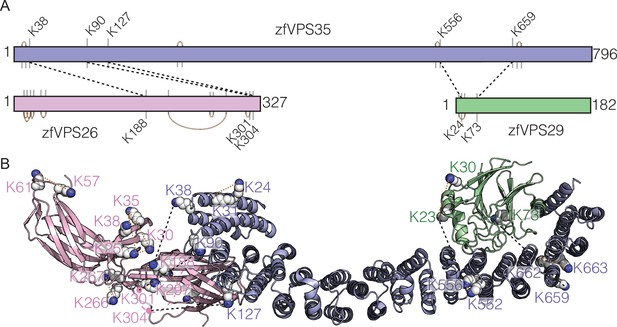
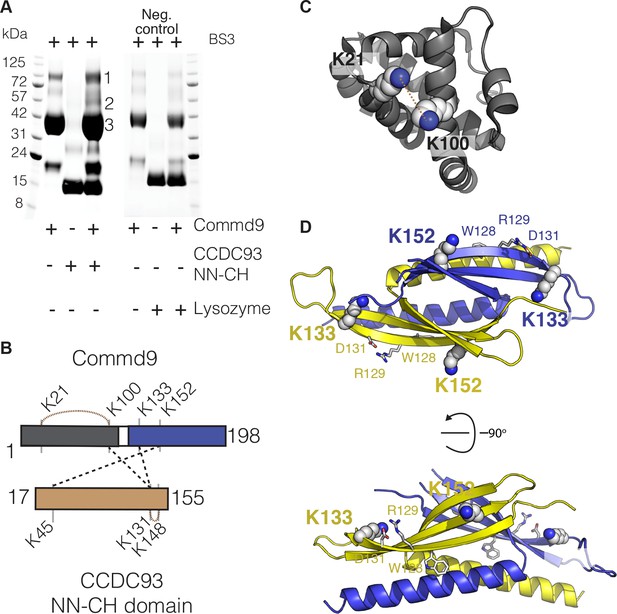
Cross-linking of zfRetromer complex.
(A, B) Cross-linked map for zfRetromer complex and the representative complex mode highlighting the cross-linked region. Intramolecular and intermolecular cross-linked peptides are labelled in the same format as Commd9 and the NN-CH domain of CCDC93 cross-links. Retromer complex model was generated using the crystal structures of VPS29 – VPS35 (PDB ID: 2R17) (Hierro et al., 2007) and VPS26 – VPS35 (PDB ID: 5F0J) (Lucas et al., 2016) as template.
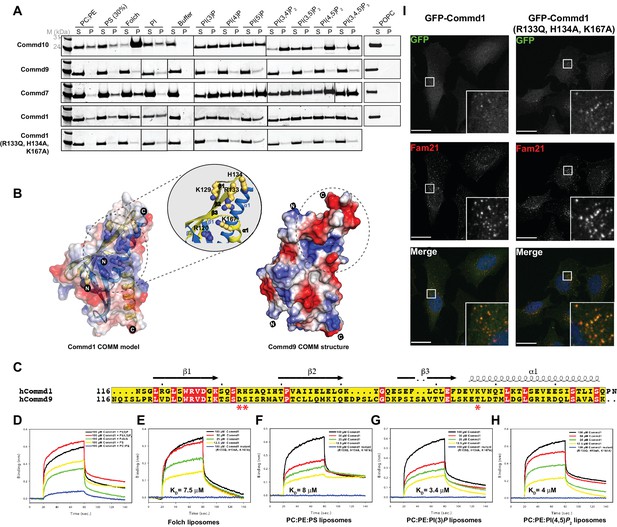
COMMD proteins bind membrane phospholipids but this is not required for endosomal recruitment.
(A) Commd1, Commd1 (R133Q, H134A, K167A), Commd7, Commd9 and Commd10 were incubated with POPC (100%), POPC/POPE (90:10, molar ratio), POPC/POPE/POPS (60:10:30, molar ratio) and POPC/POPE/PIP (80:10:10, molar ratio) liposomes doped with different phosphoinositides to perform liposome pelleting assay by ultracentrifugation and subsequent protein content analysis of the supernatant (S) and pellet (P) fractions. (B) (Left) Homology model of Commd1 COMM domain shown as a transparent electrostatic surface overlaid with the ribbon representation, highlighting the residues crucial for forming the positively charged lipid-binding pocket. Inset shows the close up of the lipid-binding pocket. (Right) The electrostatic surface representation of Commd9 COMM domain, highlighting the absence of basic patch. (C) Sequence alignment of Commd1 and Commd9 COMM domains. Red asterisks mark the positively charged amino acids constituting the basic patch on Commd1, which are absent in Commd9. (D–H) Binding of liposomes containing various phosphoinositides to Commd1 measured at different concentrations and Commd1 (R133Q, H134A, K167A) using the BLiTz system. Binding kinetics was calculated using sigma-plot (Systat Software Inc.). (I) Lentiviral constructs of GFP-tagged Commd1 and Commd1 (R133Q, H134A, K167A) were transfected into HeLa cells and colocalisation with the Fam21 WASH subunit imaged by confocal immunofluorescence microscopy. The mutant Commd1 is still recruited to endosomes, presumably due to incorporation into heteromeric COMMD complexes and the CCC/Retreiver subunits.
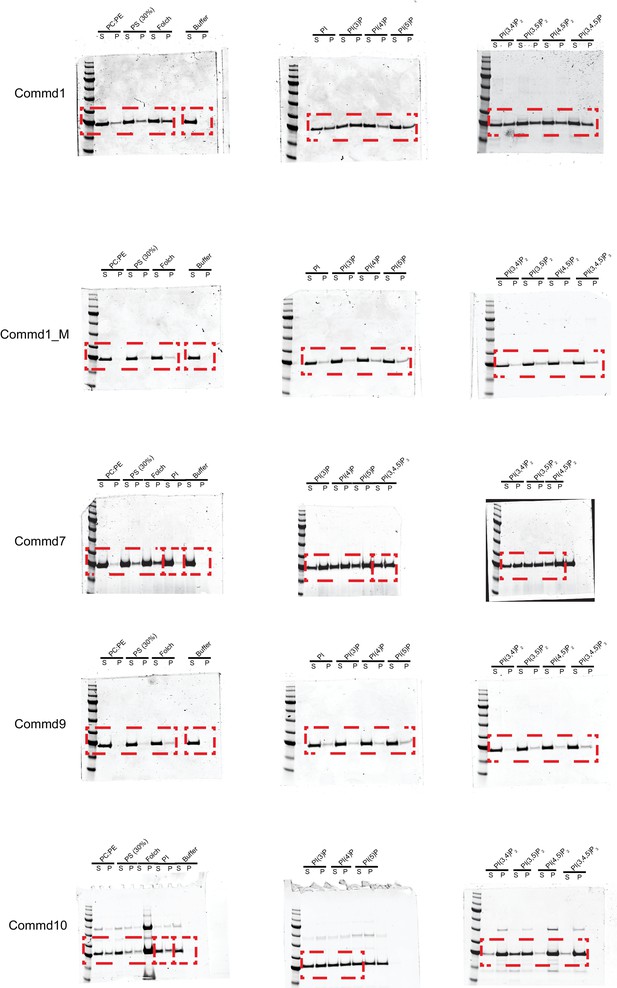
Raw files of SDS-PAGE gels of liposome pelleting assay with COMMD proteins.
https://doi.org/10.7554/eLife.35898.022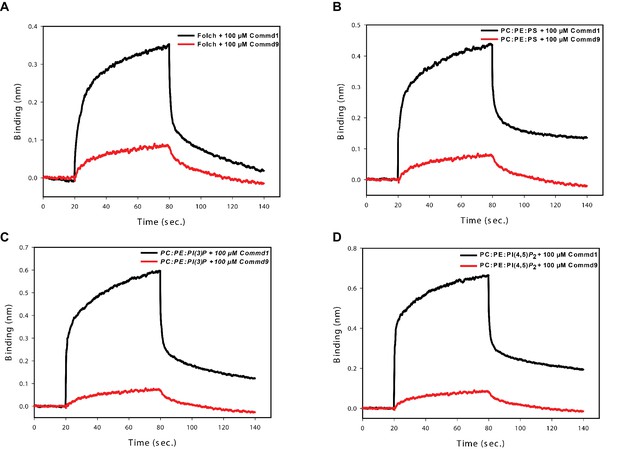
Binding comparison of Commd1 and Commd9 with liposomes using BLiTz.
https://doi.org/10.7554/eLife.35898.023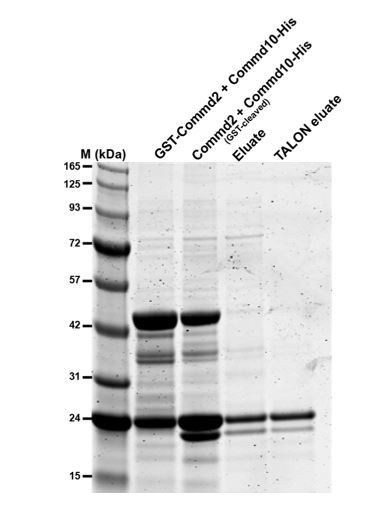
Commd2-Commd10 co-purification.
GST-Commd2 was co-expressed with Commd9-His, proteins were then purified by GST-affinity chromatography, thrombin cleavage and then His-tag affinity chromatography. The two proteins co-purify in a stable heterodimer (Commd10-His is the upper band and cleaved Commd2 is the lower band.
Tables
Summary of crystallographic structure determination statistics.*
https://doi.org/10.7554/eLife.35898.004| Commd9 HN domain (1–117) | Commd9 COMM domain (115–198) | ||
|---|---|---|---|
| Data collection | |||
| Space group | P 1 | I 4 | |
| 28.5 Å, 35.6 Å, 54.3 Å; | 79.4 Å, 79.4 Å, 58.5 Å | ||
| Unit cell dimensions (a,b,c; α,β,γ) | 104.0°, 93.3°, 91.1° | 90°, 90°, 90° | |
| Remote | Inflection | ||
| Wavelength (Å) | 0.9762 | 0.9796 | 0.9787 |
| Total reflections | 172817 | 146841 | 131332 |
| Resolution range (Å) | 19–84 – 1.55 (1.58) | 19–84 – 1.55 (1.58) | 47.13–2.17 (2.24) |
| Mean I/sigma(I) | 16.9 (5.2) | 25.2 (9.3) | 26.7 (4.9) |
| R-merge | 0.10 (0.54) | 0.06 (0.18) | 0.06 (0.5) |
| Unique reflections | 27698 | 27442 | 9689 |
| Multiplicity | 6.2 (6.0) | 5.4 (5.3) | 13.6 (11.8) |
| Anomalous Multiplicity | 3.1 (3.1) | 2.6 (2.7) | 6.9 (6.1) |
| Mn(I) half-set correlation CC(1/2) | 0.99 (0.89) | 0.99 (0.98) | 0.99 (0.95) |
| Completeness (%) | 91.9 (54.9) | 91.0 (52.7) | 99.7 (96.4) |
| Anomalous Completeness (%) | 89.6 (50.3) | 88.1 (48.1) | 99.3 (93.0) |
| Wilson B-factor | 8.4 | 18.6 | 53.1 |
| Refinement | |||
| R-work | 0.13 (0.25) | 0.24 (0.30) | |
| R-free | 0.16 (0.33) | 0.28 (0.35) | |
| Resolution range (Å) | 26.28–1.55 | 30.36–2.17 | |
| Number of atoms | 2078 | 1268 | |
| Protein atoms | 1784 | 1259 | |
| RMS(bonds) | 0.009 | 0.002 | |
| RMS(angles) | 1.158 | 0.549 | |
| Ramachandran favored (%) | 100 | 98.06 | |
| Ramachandran outliers (%) | 0 | 0 | |
| Average B-factor | 13.11 | 85.61 | |
-
*Highest resolution shell is shown in parentheses.
Summary of SAXS structural parameters.
https://doi.org/10.7554/eLife.35898.013| Commd1 | Commd7 | Commd9 | |
|---|---|---|---|
| Data collection parameters | |||
| Instrument | Australian Synchrotron SAXS/WAXS beamline with Dectris PILATUS 1M detector | ||
| Wavelength (Å) | 1.0332 | ||
| Beam geometry (μM) | 250 × 130 | ||
| Camera length (m) | 1.6 | ||
| q-range (Å−1) | 0.011–0.251 | ||
| Absolute scaling method | Comparision with scattering from 1 mm H2O | ||
| Normalization | To transmitted intensity by beam-stop counter | ||
| Method for monitoring radiation damage | Dose maintained below 210 Gy | ||
| Exposure per frame (s) | 1 | ||
| Sample temperature (K) | 283 | ||
| Sample configuration | SEC-SAXS with sheath-flow cell | ||
| Flow-rate (ml/min) | 0.25 | ||
| Structural parameters | |||
| Guinier analysis | |||
| I(0) (cm−1) | 0.04 ± 0.00 | 0.03 ± 0.00 | 0.02 ± 0.00 |
| Rg (Å) | 29.26 ± 0.25 | 28.86 ± 0.60 | 27.61 ± 0.79 |
| qmin (Å−1) | 0.01 | 0.01 | 0.01 |
| qRg max (Å−1) | 1.3 | 1.3 | 1.3 |
| P(r) analysis | |||
| I(0) (cm−1) | 0.04 ± 0.0004 | 0.029 ± 0.0003 | 0.02 |
| Rg (Å) | 30.37 ± 0.59 | 29.5 ± 0.53 | 28.26 ± 0.67 |
| Dmax (Å) | 101.47 | 99.47 | 96.99 |
| q range (Å−1) | 0.011–0.251 | 0.011–0.251 | 0.011–0.251 |
| Porod volume (Å−3) | 71951 | 65393 | 67180 |
| Dry volume calculated from sequence (Å−3) | 25848 | 27271 | 26622 |
| Shape model-fitting results | |||
| Gasbor (default paramters, 20 calculations | |||
| q range (Å−1) for fitting | 0.011–0.251 | 0.011–0.251 | 0.011–0.251 |
| Symmetry, anisotropy assumptions | P2, none | P2, none | P2, none |
| NSD (standard deviation) | 0.16 (0.004) | 1.04 (0.55) | |
| χ2 range, CORMAP P values | 0.15–0.20, 0.11 | 0.14–0.17, 0.47 | 0.14–0.15, 0.14 |
| Atomistic modelling | |||
| Crystal structures | PDB entry 2H2M and 6BP6 | PDB entry 4OE9 and 6BP6 | PDB entry 4OE9 and 6BP6 |
| q range for all modelling | 0.011–0.251 | 0.011–0.251 | 0.011–0.251 |
| CRYSOL (with default parameters) | |||
| No constant subtraction | |||
| χ2 | 0.31 | 0.72 | 1.14 |
| Predicted Rg (Å) | 30.85 | 30.54 | 31.88 |
| Vol (Å), Ra (Å), Dro (eÅ−3) | 53643, 1.40, 0.007 | 46010, 1.400, 0.075 | 57201, 1.400, 0.007 |
| FoXS | |||
| χ2, CORMAP P values | 0.52, 0.99 | 0.78, 0.99 | 0.99, 0.93 |
| Predicted Rg (Å) | 30.82 | 29.18 | 31.84 |
| c1,c2 | 1.03, 0.29 | 1.05, 4.0 | 1.04, 1.20 |
| Molecular mass determination | |||
| Estimated MW from Porod volume | 41387 Da | 38094 Da | 39099 Da |
| Calculated MW from MALLS | 40400 Da | 53000 Da | 47800 Da |
| Calculated MW from sequence | 21362 Da (42724 Da for dimer) | 22540 Da (45080 Da for dimer) | 21819 Da (43639 Da for dimer) |
| Software employed | |||
| Primary data reduction | Australian Synchrotron SAXS/WAXS data reduction package (Scatterbrain) | ||
| Data Processing | PRIMUS and GNOM | ||
| Ab initio modeling | GASBOR | ||
| Validation and averaging | DAMAVER | ||
| Computation of models | SASREF | ||
| Envelope representations | PyMOL | ||
Identified cross-linked peptides of Commd9 and NN-CH domain of CCDC93.
https://doi.org/10.7554/eLife.35898.019| Predicted mass (Da) | Observed mass (Da) | Error (ppm) | xQuest score | ||
|---|---|---|---|---|---|
| Cross-linked peptides | |||||
| Sample 1: | Commd9 - Commd9 | ||||
| 2751.419 | 2751.407 | 2.3 | 21.10 | VDIK(133)TSSDSISR-VDIK(133)TSSDSISR | |
| 2305.209 | 2305.206 | 3.0 | 20.35 | ASSK(21)DVVR-VDIK(133)TSSDSISR | |
| 4316.319 | 4316.306 | 3.2 | 18.78 | DLSSAEAILALFPENFHQNLK(95)NLLTK(100)IILEHVSTWR | |
| 3266.789 | 3266.782 | 1.8 | 18.67 | NLLTK(100)IILEHVSTWR-VDIK(133)TSSDSISR | |
| 2820.589 | 2820.579 | 2.4 | 17.28 | ASSK(21)DVVR-NLLTK(100)IILEHVSTWR | |
| 3648.889 | 3648.881 | 1.0 | 15.22 | LVDLDWRVDIK()TSSDSISR-VDIK(133)TSSDSISR | |
| 3202.679 | 3202.677 | 2.0 | 14.22 | ASSK(21)DVVR-LVDLDWRVDIK(133)TSSDSISR | |
| 2571.299 | 2571.288 | 4.1 | 12.78 | IQEDPSLCGDK(163)PSISAVTVELSK(175) | |
| 1859.009 | 1859.006 | 2.7 | 12.40 | ASSK(21)DVVR-ASSK(21)DVVR | |
| Sample 2: | Commd9 - CCDC93 | ||||
| 2998.479 | 2998.476 | 1.5 | 16.86 | VDIK(133)TSSDSISR | AIETK(131)EEMGDYIR |
| 3513.859 | 3513.842 | 1.4 | 14.74 | NLLTK(100)IILEHVSTWR | AIETK(131)EEMGDYIR |
| 4772.479 | 4772.501 | 5.1 | 7.11 | MAVPTCLLQMK(152)IQEDPSLCGDKPSISAVTVELSK | IK(45)GLSPFDK |
| Commd9 - Commd9 | |||||
| 2820.589 | 2820.583 | 1.1 | 18.60 | ASSK(21)DVVR-NLLTK(100)IILEHVSTWR | |
| CCDC93 - CCDC93 | |||||
| 3569.679 | 3569.675 | 0.3 | 14.92 | AIETK(131)EEMGDYIR-SYSVSQFQK(148)TYSLPED | |
| Sample 3: | Commd9 - Commd9 | ||||
| 2820.589 | 2820.599 | 4.6 | 20.02 | ASSK(21)DVVR-NLLTK(100)IILEHVSTWR | |
| 2305.209 | 2305.226 | 5.8 | 19.51 | ASSK(21)DVVR-VDIK(133)TSSDSISR | |
Identified cross-linked peptides of zfRetromer.
https://doi.org/10.7554/eLife.35898.020| Predicted mass (Da) | Observed mass (Da) | Error (ppm) | xQuest score | |||
|---|---|---|---|---|---|---|
| Intermolecular cross-linked peptides | zfVPS26 | zfVPS29 | zfVPS35 | |||
| 2929.569 | 2929.550 | 8.0 | 13.20 | APEK(301)MR | K(90)VADLYELVQYAGNIIPR | |
| 1844.019 | 1844.040 | 9.4 | 10.12 | SKYHLK(188) | NK(38)LMDALK | |
| 1342.809 | 1342.805 | 4.9 | 9.99 | MRK(304)R | K(127)DILK | |
| 1756.069 | 1756.070 | 2.8 | 9.83 | FK(23)KLLVPGK | WEKK(556) | |
| 3809.959 | 3809.982 | 7.1 | 8.01 | GDFDENLNYPEQK(73)VVTVGQFK | TQCALAASK(659)LLK | |
| Intramolecular cross-linked peptides | zfVPS26 – zfVPS26 | |||||
| 1441.829 | 1441.819 | 3.3 | 28.12 | VNINVK(57)QTSK(61)R | ||
| 1630.869 | 1630.863 | 4.6 | 26.54 | TAELK(30)TEEGK(35)LEK | ||
| 1247.689 | 1247.679 | 2.1 | 26.15 | DVNK(266)K(267)FSVR | ||
| 2571.239 | 2571.229 | 4.3 | 22.06 | TEEGK(35)LEK(38)HYLFYDGESVSGK | ||
| 3241.639 | 3241.662 | 7.0 | 20.14 | K(25)TAELKTEEGK(35)LEKHYLFYDGESVSGK | ||
| 3626.859 | 3626.875 | 5.8 | 19.78 | K(214)EMTGIGPSTTTETETVAK-YFK(288)QQEIVLWR | ||
| 844.489 | 844.487 | 4.4 | 18.66 | K(25)TAELK(30) | ||
| 1014.549 | 1014.544 | 1.4 | 12.32 | K(297)APEK(301)MR | ||
| Intramolecular cross-linked peptides | zfVPS29 - zfVPS29 | |||||
| 1184.749 | 1184.736 | 5.9 | 18.59 | FK(23)KLLVPGK(30) | ||
| Intramolecular cross-linked peptides | zfVPS35 – zfVPS35 | |||||
| 1607.729 | 1607.726 | 3.3 | 22.38 | ENSSSDDK(552)WEKK(556) | ||
| 844.469 | 844.449 | 6.9 | 19.26 | EK(208)REK(211) | ||
| 2245.219 | 2245.215 | 5.1 | 18.55 | LLDEAVQAVK(24)VQSFQMK(31)R | ||
| 1255.699 | 1255.688 | 2.5 | 17.69 | LLK(662)K(663)PDQCR | ||
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.35898.024





