Differential expression of Lutheran/BCAM regulates biliary tissue remodeling in ductular reaction during liver regeneration
Figures
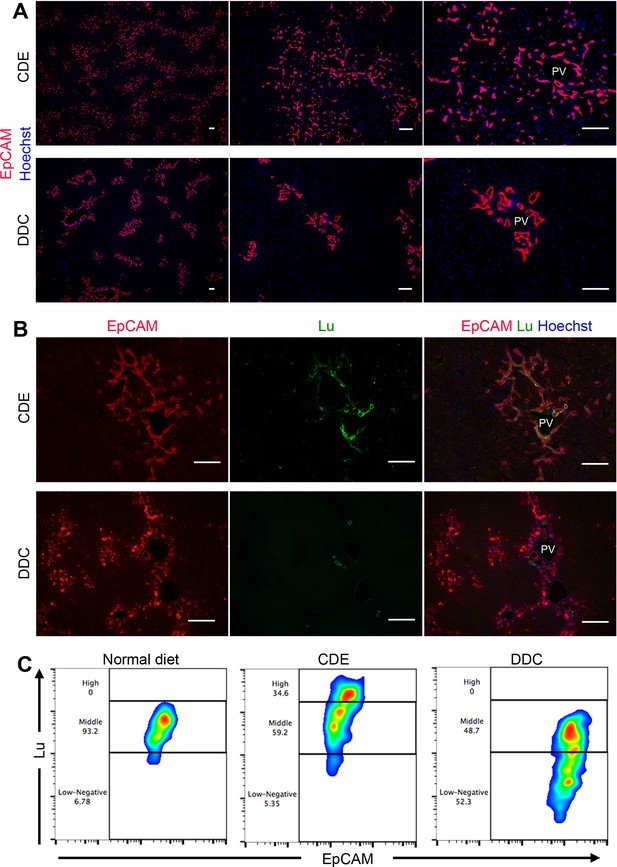
Phenotypic difference of DR and biliary cells between the CDE model and DDC model.
(A) Immunohistochemical analysis of CDE-fed and DDC-fed mouse liver sections for EpCAM. (B) Co-localization of EpCAM and Lu in CDE-fed and DDC-fed mouse liver sections. (C) Comparison of Lu expression level in EpCAM+ cells among normal diet-fed, CDE-fed and DDC-fed mouse livers by flow cytometric analysis. PV: portal vein. Scale bar: 100 μm.
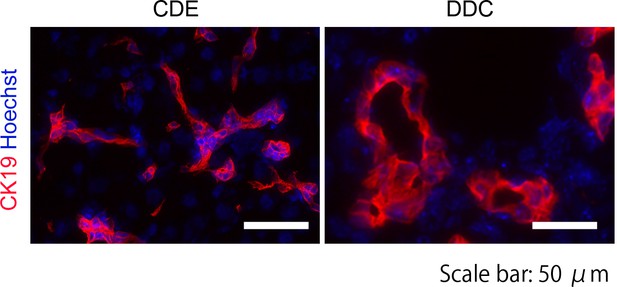
Immunohistochemical analysis of CDE-fed and DDC-fed mouse liver sections with anti-CK19 antibody.
The LPCs in the CDE model exhibit spindle like shape forming primitive ductules, while those in the DDC model show obvious duct structure. Scale bar: 50 μm.
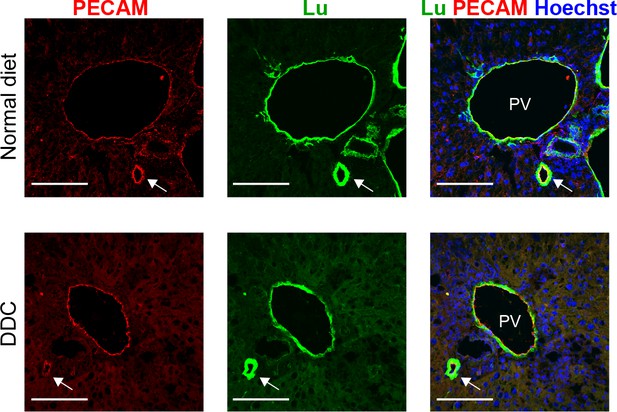
Expression analysis for PECAM and Lu in normal and injured liver.
Co-staining of PECAM and Lu was performed in liver sections of normal diet- and DDC-fed mouse liver. Arrows show a doubly stained hepatic artery. PV: portal vein. Scale bar: 100 μm.
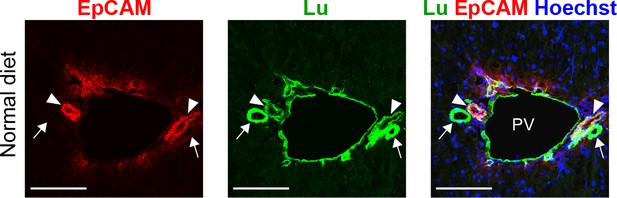
Co-staining of EpCAM and Lu in liver sections of normal liver.
Arrows and arrowheads point to hepatic arteries and bile ducts, respectively. The fluorescence signal of Lu is weakly detected in EpCAM+ bile ducts, while it is saturated in EpCAM- hepatic arteries in this setting of exposure. PV: portal vein. Scale bar: 100 μm.
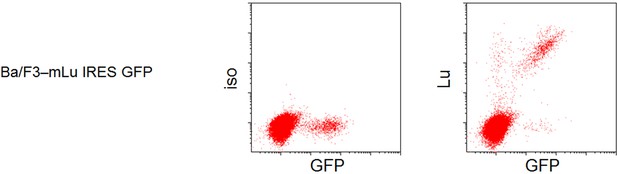
Validation of specific reactivity of the used antibody to Lutheran.
Ba/F3 cell, a murine pro B-cell line, was transduced with retroviral vector containing cDNA encoding mouse Lu and green fluorescent protein (GFP). By FCM analysis, the only GPF-positive cells that express Lu shift with anti-Lu antibody.
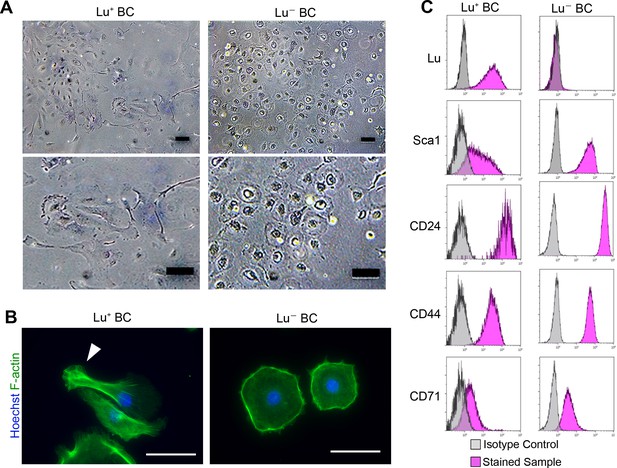
Culture of Lu+and Lu- BC isolated from injured liver.
(A) Representative images of Lu+ BC and Lu- BC by bright field microscopy. (B) Immunocytochemistry for F-actin in cultured Lu+ BC and Lu- BC. Arrowhead indicates pseudopod. (C) Flow cytometric analysis of Lu, Sca1, CD24, CD44, and CD71 expression levels in Lu+ BC and Lu- BC. The cultured Lu+ BC and Lu- BC were used for analysis after 6 passages. Scale bar: 100 μm.
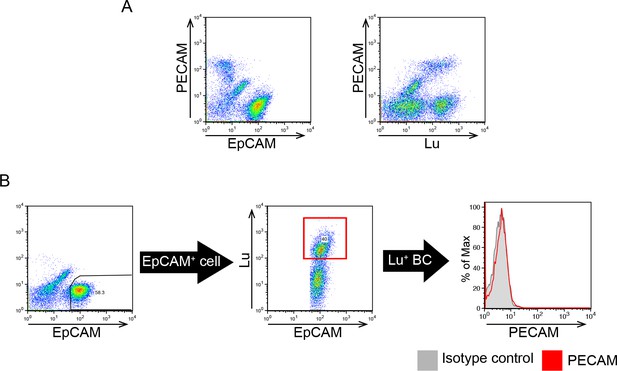
Expression profile of EpCAM, Lu and PECAM in non-parenchymal cells (NPCs) prepared from CDE-injured livers.
NPCs were analyzed by flow cytometry after triple staining with anti-EpCAM, anti-Lu and anti-PECAM antibodies. (A) FCM analysis of EpCAM, Lu and PECAM expression. EpCAM+ fraction is clearly distinguished from PECAM+ fraction by FACS. (B) Gating strategy for the isolation of EpCAM+ cells by FACS. There is no contamination of PECAM+ cells in the gate of EpCAM+Lu+ biliary cells.
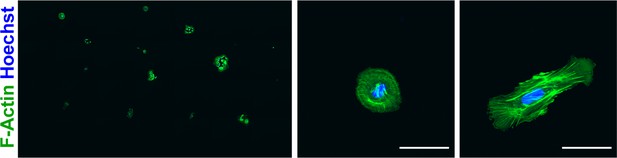
Immunocytochemistry for F-actin in cultured EpCAM+cells isolated from normal liver.
The image was acquired 7 days after primary culture. The attached cells showed a mixture of round and indefinite shape. Scale bar: 100 μm.
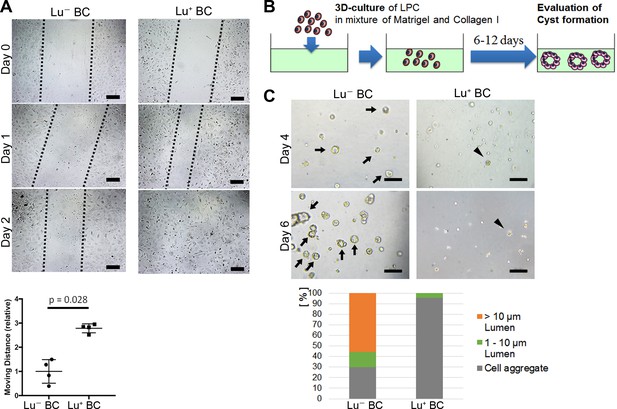
Evaluation of Lu- BC and Lu+ BC characteristics by scratch assay and cyst formation assay.
(A) Scratch assay using Lu- BC or Lu+ BC. Representative images of day 0, day 1, and day 2 after scratch are shown. Quantitative data of cell moving distance at day 1 are plotted in a graph with mean and standard deviation. n = 4 biological replicates. Dotted line indicates cell front of scratched gap. (B) Schematic diagram of three-dimensional culture and cyst generation. (C) Bright field microscopic image of three-dimensionally cultured Lu+ BC and Lu- BC at culture day 4 or day 6. Arrows and arrowheads point to cysts and cell aggregates, respectively. The details of formed cysts are shown below. The cell cluster devoid of luminal structure was regarded as ‘Cell aggregate’. Scale bar: 100 μm.
-
Figure 3—source data 1
Figure 3A: Numerical data for measurements of migrating distance in the scratch assay using Lu- BC and Lu+ BC.
Figure 3C: Numerical data for the details of formed cyst in the 3D culture using Lu- BC and Lu+ BC.
- https://doi.org/10.7554/eLife.36572.012
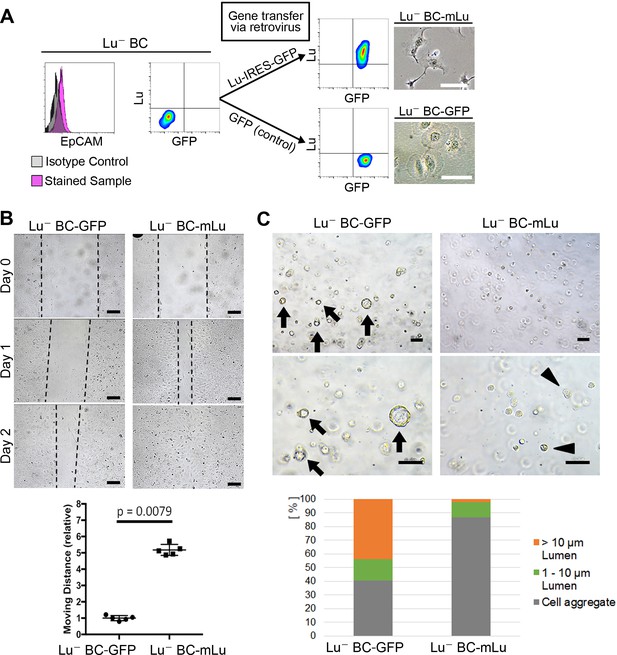
Lu regulates the motility and cyst formation capacities of biliary cells.
(A) Schematic description for generating Lu- BC-mLu or Lu- BC-GFP by using retrovirus vector. Flow cytometric analyses show expression level of Lu and GFP. Bright field microscopic images show morphological change in Lu- BC-mLu. (B) Scratch assay using Lu- BC-GFP or Lu- BC-mLu. Representative images of day 0, day 1, and day 2 after scratch are shown. Quantitative data of cell moving distance at day 1 are plotted in a graph with mean and standard deviation. n = 5 biological replicates. Dotted line indicates cell front of scratched gap. (C) Bright field microscopic image of three-dimensionally cultured Lu- BC-GFP or Lu- BC-mLu at culture day 6. Arrows and arrowheads point to cysts and cell aggregates, respectively. The details of formed cysts are shown below. The cell cluster devoid of luminal structure was regarded as ‘Cell aggregate’. Scale bar: 100 μm.
-
Figure 4—source data 1
Figure 4B: Numerical data for measurements of migrating distance in the scratch assay using Lu- BC-GFP and Lu- BC-mLu.
Figure 4C: Numerical data for the details of formed cyst in the 3D culture using Lu- BC-GFP and Lu- BC-mLu.
- https://doi.org/10.7554/eLife.36572.014
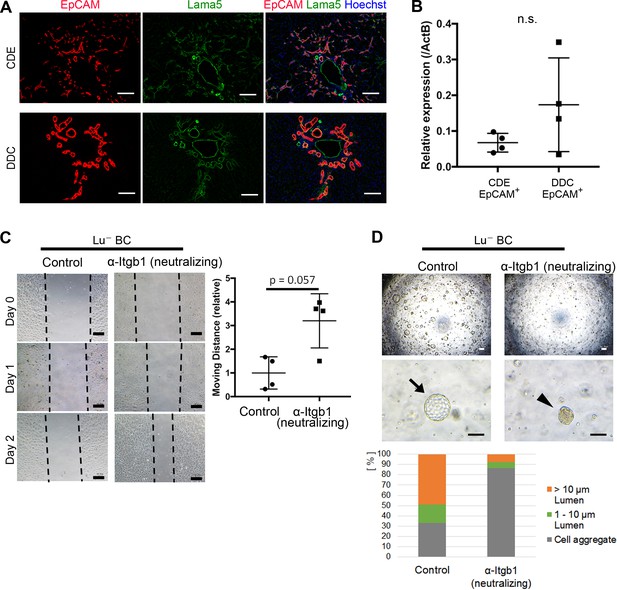
Itgb1 signaling is critical for regulating the phenotype of biliary cells.
(A) Expression analysis for Lama5 in injured liver. Co-staining of EpCAM and Lama5 was performed in liver sections of CDE-fed mouse and DDC-fed mouse. (B) Evaluation of Lama5 gene expression in EpCAM+ cells isolated from CDE-fed and DDC-fed mouse livers by quantitative RT-PCR. Data are plotted in a graph with mean and standard deviation. n = 4 biological replicates. n.s.: not significance. (C) Scratch assay using Lu- BC in the presence or absence of Itgb1 neutralizing antibody. Hamster IgM was used as control. Representative images of day 0, day 1, and day 2 after scratch are shown. Dotted line indicates cell front of scratched gap. Quantitative data of cell moving distance at day 1 are plotted in a graph with mean and standard deviation. n = 4 biological replicates. (D) Bright field microscopic image of three-dimensionally cultured Lu- BC-GFP at culture day 12 in the presence of Itgb1 neutralizing antibody or control IgM. Arrows and arrowheads point to cysts and cell aggregates, respectively. The details of formed cysts are shown below. The cell cluster devoid of luminal structure was regarded as ‘Cell aggregate’. Scale bar: 100 μm.
-
Figure 5—source data 1
Figure 5B: Numerical data for expression analysis of Lama5 mRNA in EpCAM+ cells by quantitative RT-PCR.
Figure 5C: Numerical data for measurements of migrating distance in the scratch assay using Lu- BC in the presence or absence of neutralizing anti-Itgb1 antibody. Figure 5D: Numerical data for the details of formed cyst in the 3D culture using Lu- BC in the presence or absence of neutralizing anti-Itgb1 antibody.
- https://doi.org/10.7554/eLife.36572.023

Schematic model of the inhibitory effect of Lutheran on Integrin signaling.
Lama5-containing laminins are able to bind to both Lutheran and Integrinα3β1/α6β1. High expression of Lutheran in LPC inhibits β1 Integrin signaling mediated by Laminin-511/521 competitively.
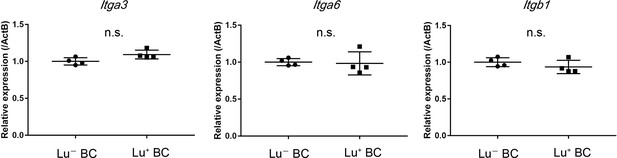
Gene expression analysis of Itga3, Itga6 and Itgb1 mRNA in Lu- and Lu+ BC by quantitative RT-PCR.
Relative expression level is plotted in a graph with mean and standard deviation (n = 4). n.s.: not significance.
-
Figure 5—figure supplement 2—source data 1
Numerical data for expression analysis of Itga3, Itga6 and Itgb1 mRNA in Lu+ BC and Lu- BC by quantitative RT-PCR.
- https://doi.org/10.7554/eLife.36572.018
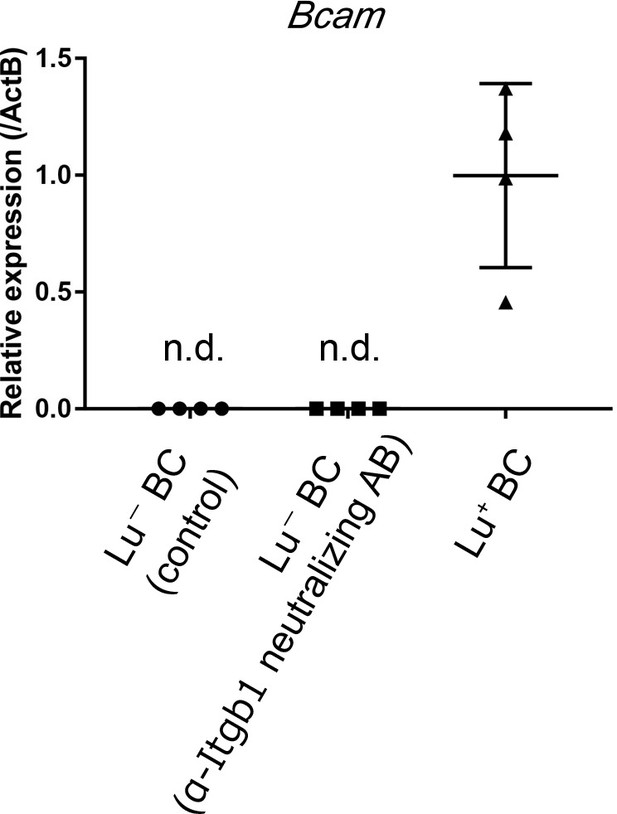
Gene expression analysis of Bcam mRNA in Lu- BC.
Lu- BC cultured in the presence or absence of neutralizing anti-Itgb1 antibody was analyzed by quantitative RT-PCR. Lu+ BC was used as a positive control. Relative expression level is plotted in a graph with mean and standard deviation (n = 4). n.d.: not detected.
-
Figure 5—figure supplement 3—source data 1
Numerical data for expression analysis of Bcam mRNA in Lu- BC by quantitative RT-PCR.
- https://doi.org/10.7554/eLife.36572.020
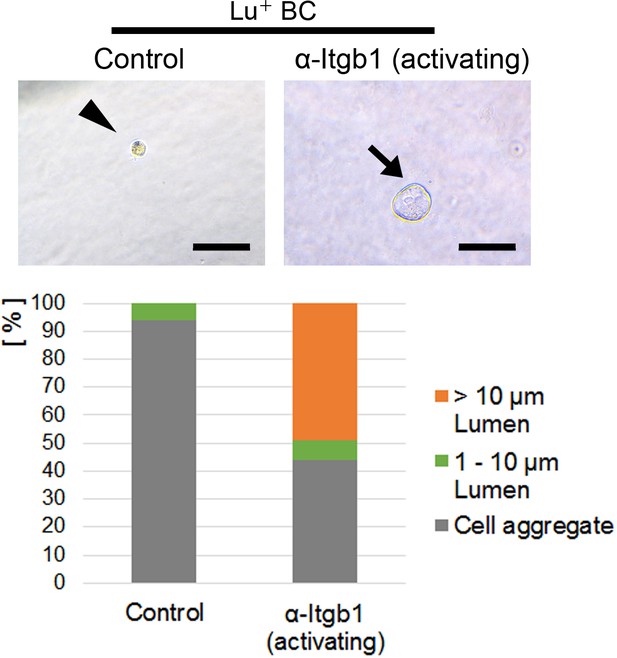
Cyst formation assay of Lu+ BC in the presence of activating anti-Itgb1 antibody (TS2/16) or control IgM.
Representative images of cultured Lu+ BC at culture day 6 are shown. Arrow and arrowhead point to a cyst and a cell aggregate, respectively. The details of formed cysts are shown below. The cell cluster devoid of luminal structure was regarded as ‘Cell aggregate’. Scale bar: 100 μm.
-
Figure 5—figure supplement 4—source data 1
Numerical data for the details of formed cyst in the 3D culture using Lu+ BC in the presence or absence of activating anti-Itgb1 antibody (TS2/16).
- https://doi.org/10.7554/eLife.36572.022
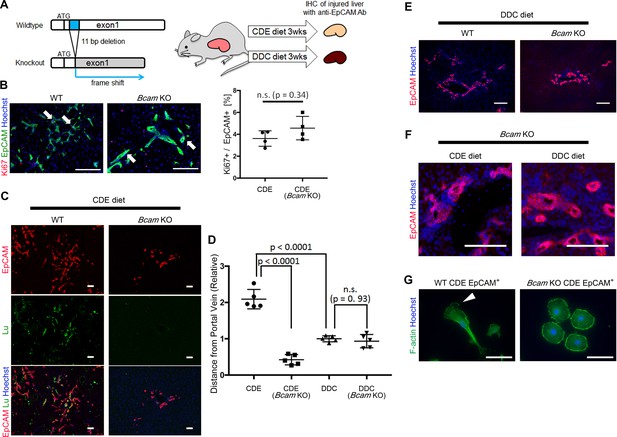
Bcam KO mice show drastic phenotype change in DR.
(A) Schematic diagram of experiments using Bcam KO mouse. (B) Co-staining of EpCAM and Ki67 in WT and Bcam KO mouse liver sections in the CDE model. The ratio of Ki67+ cell per EpCAM+ cell is plotted in a graph with mean and standard deviation. n = 4 biological replicates. n.s.: not significance. (C) Co-staining of EpCAM and Lu in CDE-fed WT and Bcam KO mouse liver sections. (D) Quantitative analysis for the distance from portal vein to distal biliary cells in the CDE and DDC models. Data are plotted in a graph with mean and standard deviation. Statistical significance among groups is determined using one-way ANOVA. n = 5 biological replicates. (E) Immunostaining of EpCAM in WT and Bcam KO mouse liver sections in the DDC model. (F) Magnified immunohistochemical image of DR in CDE-fed and DDC-fed Bcam KO mouse liver. (G) Morphological image of EpCAM+ cells sorted from WT and Bcam KO mouse fed a CDE diet for 3 weeks. The cells were directly stained with Phalloidin to visualize F-actin 72 hr after plating on culture dish. Arrowhead points to pseudopod. Scale bar: 100 μm.
-
Figure 6—source data 1
Figure 6B: Numerical data for the ratio of Ki67+ cells per EpCAM+ cells.
Figure 6D: Numerical data for measurements of the distance from portal vein to distal biliary cells in the CDE and DDC models.
- https://doi.org/10.7554/eLife.36572.026
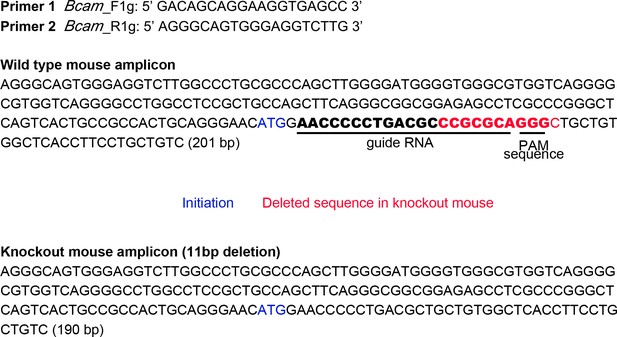
Generation of Bcam KO mouse by using the CRISPR/Cas9 method.
Schematic illustration of the mouse Lu/Bcam gene structure and sequences around the targeted allele. Blue and red letters stand for an initiation ATG codon and the deleted sequence (11 bp) in KO mouse, respectively. The sequence for gRNA and protospacer-adjacent motif (PAM) are underlined. Genomic PCR of WT and Bcam KO mice using a set of Primer 1 and Primer 2 yields amplicon corresponding to 201 and 190 bp, respectively. Information on Source data.
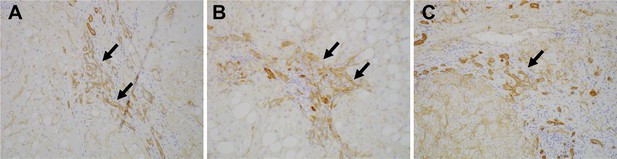
Expression of CD239 in human liver disease.
Immunohistochemical images for Lu/BCAM (CD239) in human liver disease are shown. Cirrhotic liver sections obtained from the patients of non-alcoholic steatohepatitis (A), hepatitis B virus (B) and hepatitis C virus (C) were stained. DRs are denoted with arrows.
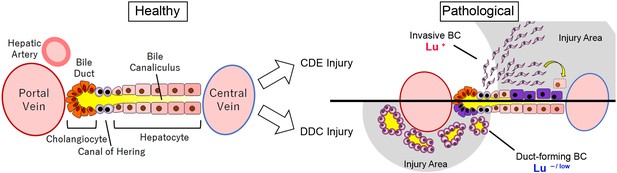
Graphical abstract of this study.
https://doi.org/10.7554/eLife.36572.028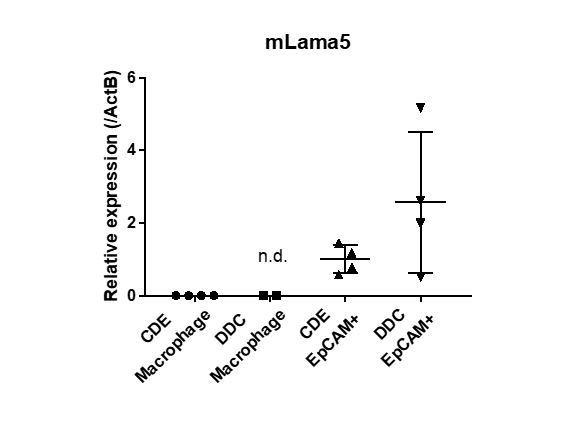
Tables
| Reagent type | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-EpCAM (rat monoclonal) | PMID: 19429791 | (1:100–500) | |
| Antibody | anti-EpCAM (rabbit polyclonal) | Abcam | Abcam:ab71916; RRID:AB_1603782 | (1:400) |
| Antibody | anti-Lutheran (rat monoclonal) | this paper | (1:500) | |
| Antibody | anti-Sca1 (rat monoclonal) | BioLegend | BioLegend:108107; RRID:AB_313344 | (1:400) |
| Antibody | anti-CD24 (rat monoclonal) | Miltenyi Biotec | MB:130-102-731; RRID:AB_2656573 | (1:400) |
| Antibody | anti-CD71 (rat monoclonal) | Miltenyi Biotec | MB:130-109-632; RRID:AB_2659126 | (1:400) |
| Antibody | anti-CD44 (rat monoclonal) | Miltenyi Biotec | MB:130-110-117; RRID:AB_2658152 | (1:400) |
| Antibody | anti-FcR (rat monoclonal) | BioLegend | BioLegend:101320; RRID:AB_1574975 and BioLegend:101302; RRID:AB_312801 | (1:100) |
| Antibody | anti-CK19 (rabbit polyclonal) | PMID:12665558 | (1:1000) | |
| Antibody | anti-Lama5 (rabbit polyclonal) | PMID:9151674 | (1:200) | |
| Antibody | anti-Ki67 (rat monoclonal) | ThermoFisher | ThermoFisher:14-5698-80; RRID:AB_10853185 | (1:200) |
| Antibody | anti-PECAM (rat monoclonal) | BD Biosciences | BD:553373; RRID:AB_394819 | (1:100) |
| Antibody | anti-CD31 (rabbit polyclonal) | Novus | NB:NB100-2284; RRID:AB_10002513 | (1:100) |
| Antibody | anti-CD239 (rabbit monoclonal) | Abcam | Abcam:2994–1; RRID:AB_2065309 | (1:100) |
| Antibody | anti-CD29 (hamster monoclonal) | BD Biosciences | BD:555002; RRID:AB_395636 | 5 μg/mL for 3D culture |
| Antibody | anti-CD29 (mouse monoclonal) | BioLegend | Biolegend:303010; RRID:AB_314326 | 50 μg/mL for 3D culture |
| Antibody | Hamster IgM, λ1 isotype (hamster monoclonal) | BD Biosciences | BD:553957; RRID:AB_479639 | 5 μg/mL for 3D culture |
| Antibody | Mouse IgG1, κsotype (mouse monoclonal) | BioLegend | BioLegend:401404 | 50 μg/mL for 3D culture |
| Cell line (Homo sapiens) | Plat-E (Platinum-E) | PMID: 10871756 | RRID:CVCL_B488 | Cell line established in T. Kitamura lab |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36572.029





