Essential metabolism for a minimal cell
Figures
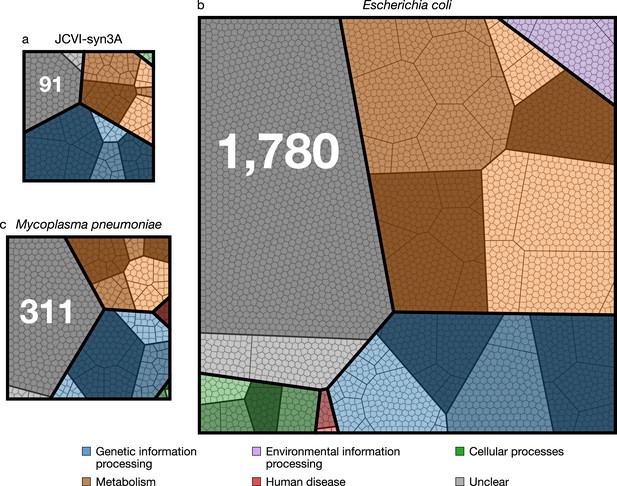
Comparison of protein coding genes in the genomes of JCVI-syn3A (NCBI GenBank: https://www.ncbi.nlm.nih.gov/nuccore/CP016816.2 (Glass, 2017)), M. pneumoniae (NCBI GenBank: https://www.ncbi.nlm.nih.gov/nuccore/U00089.2 (Himmelreich et al., 2014)), and E. coli (NCBI GenBank: https://www.ncbi.nlm.nih.gov/nuccore/NC_012967.1 (Jeong et al., 2017)) with 452, 688, and 4637 coding genes, respectively.
Each color represents a primary functional class, each contiguous shaded region corresponds to a secondary functional class, within each of the shaded regions the bold lines separate tertiary functional classes, finally each polygonal cell represents a single gene. The functional class hierarchy is presented in Supplementary file 1A. The ratio of metabolic to genetic information processing genes—0.67, 0.79, and 2.23 respectively—is smallest for JCVI-syn3A. The JCVI-syn3A genome contains both the smallest absolute number of genes of unclear function and the smallest percentage, 91 (20 %), compared to M. pneumoniae with 311 (45 %) and E. coli with 1780 (38 %).
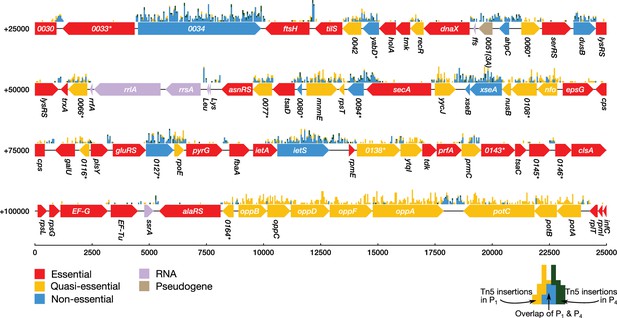
Classification of gene essentiality from transposon insertion data using a Poisson mixture model for a representative region of the JCVI-syn3A genome.
Coding regions are colored by their predicted class: red (essential), yellow (quasi-essential), blue (non-essential). Lavender regions denote RNA and light brown regions are pseudogenes. The distributions of transposon insertions in passage 1 and passage 4 are represented by yellow and dark green histograms, respectively (bin size of 50 bp). The overlap of the two histograms is highlighted in blue. When a common gene name is not available, the four-digit locus tag for JCVI-syn1.0 is used instead. Locus number identifiers with the (3A) suffix represent newly identified open reading frames in JCVI-syn3A which are missing from the JCVI-syn1.0 annotation. Asterisks mark genes with unknown functionality.
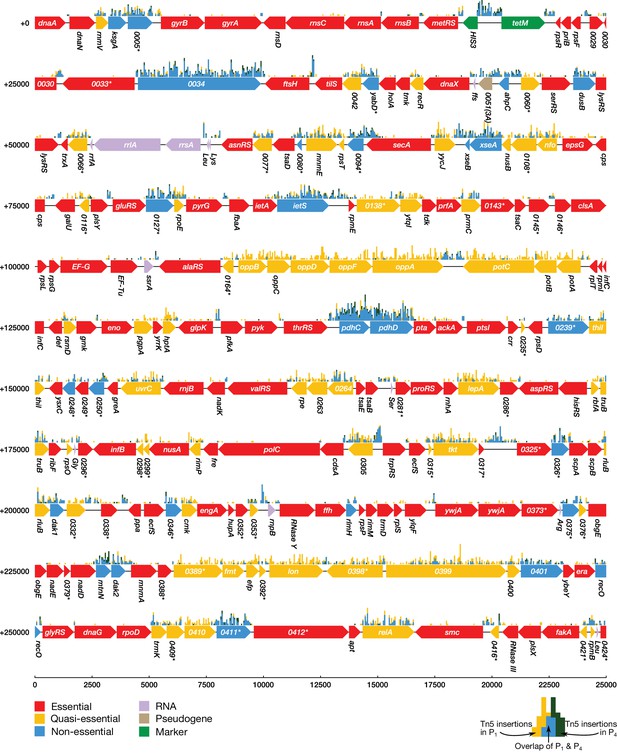
Classification of gene essentiality from transposon insertion data using a Poisson mixture model for 0–275,000 bp.
Coding regions are colored by their predicted class: red (essential), yellow (quasi-essential), blue (non-essential). Lavender regions denote RNA, light brown regions are pseudogenes, and green regions are markers used to construct and implant the genome. The distributions of transposon insertions in passage 1 and passage 4 are represented by yellow and dark green histograms respectively (bin size of 50 bp). The overlap of the two histograms is highlighted in blue. When a common gene name is not available, the four-digit locus tag for JCVI-syn1.0 is used instead. Locus number identifiers with the (3A) suffix represent represent newly identified open reading frames in JCVI-syn3A which are missing from the JCVI-syn1.0 annotation. Asterisks mark genes with unknown functionality.
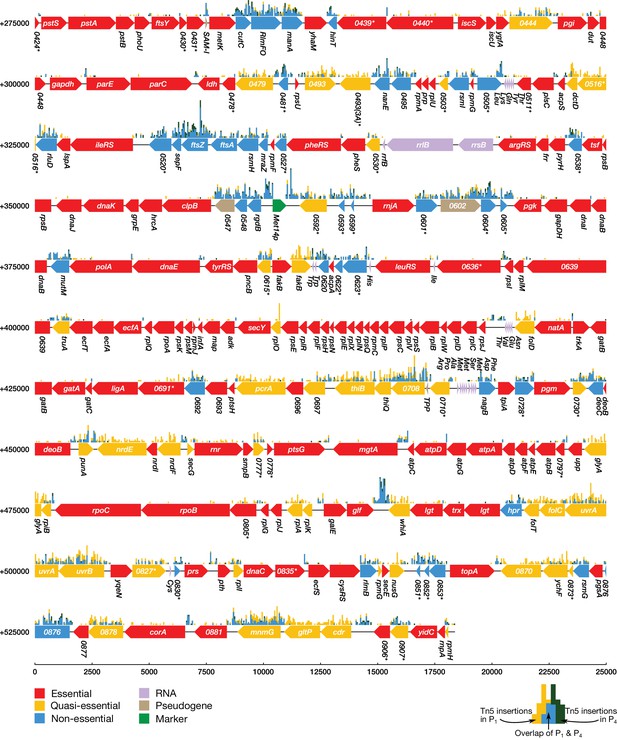
Classification of gene essentiality from transposon insertion data using a Poisson mixture model for 275,000–543,379 bp.
https://doi.org/10.7554/eLife.36842.006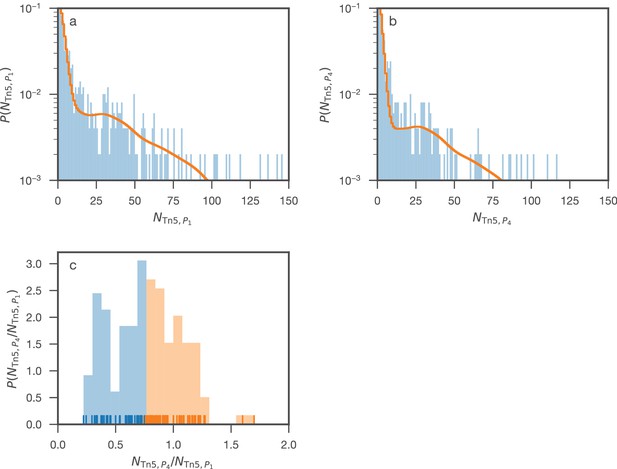
Distribution of transposon insertion counts for (panel a) and (panel b) compared to the distribution inferred through the Poisson mixture model.
To separate genes labeled ‘non-essential’ by the mixture model, but that showed a significant decrease in insertion counts from to , -means clustering was used on the ratios of transposon insertion rates in and for the genes labeled ‘non-essential’. Panel c shows how the genes were divided into two clusters such that the first cluster (blue) contains quasi-essential genes and the second contains truly non-essential genes.
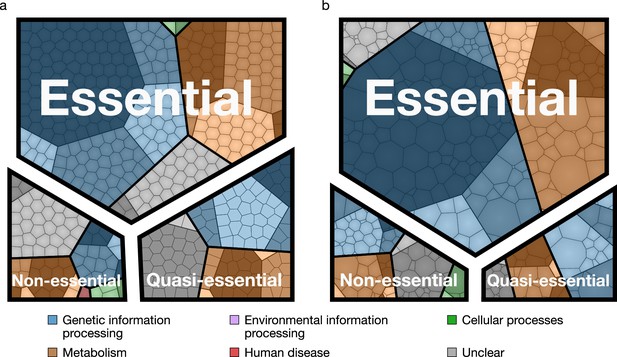
Essential, quasi-essential, and non-essential protein coding genes in JCVI-syn3A across four functional classes.
(a) Distribution across genome (cell areas all equal). (b) Distribution across proteome (cell areas proportional to protein copy number in an average cell). Among non-essential proteins, the three most abundant ones are ftsZ/0522, the peptidase 0305 and 0538 (unclear function). A detailed breakdown of the JCVI-syn3A genome into these classes is available in Table 1.

Biomass reaction equation for JCVI-syn3A.
This reaction consumes biomass precursors (macromolecules, lipids, capsule, small molecules) (black) and consumes energy in the form of ATP (red) to produce biomass (blue). Values in parentheses are the stoichiometric coefficients in mmol compound per gram cellular dry weight (mmol gDW−1). The macromolecular compositions are highlighted in green (stoichiometric coefficients within the macromolecule, unitless) and the compositions of lipids and small molecule pools are highlighted in gray (mmol gDW−1). ATP expenses within green boxes denote total macromolecular synthesis costs (based on macromolecular fractions in the biomass) and the ATP expense in the main equation denotes the nonquantifiable part of the growth-associated maintenance cost (GAM; see Section 'GAM/NGAM').
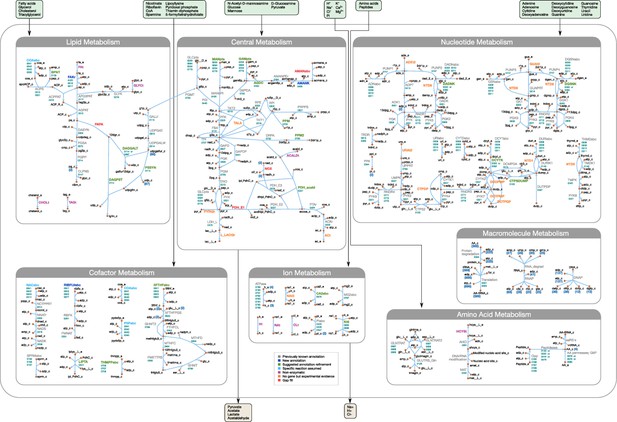
Overview of the metabolic reconstruction of JCVI-syn3A, drawn with Escher (King et al., 2015).
Orange nodes represent metabolites, labeled by their short names in the model (black); the suffixes ‘_c’ and ‘_e’ denote cytoplasmic and extracellular compartments, respectively. For clarity, H2O, H+, PPi and Pi are generally omitted as reactants. Blue edges represent (enzymatic or spontaneous) reactions, labeled by reaction name (gray labels) and associated gene loci (gene-protein-reaction (GPR) rules, turquoise; omitting ‘MMSYN1_’ prefix). Blue parenthesized numbers denote reactants (products) which are consumed (produced) in stoichiometric quantities greater than one. In this map and subsequent maps, the following color scheme for highlighted reactions is used—blue: reaction based on new annotation, light green: reaction based on suggested annotation refinement, cyan: specific reaction assumed for generic annotation, light violet: non-enzymatic reaction, orange: reaction not accounted for by gene yet but supported by experimental evidence, and red: reaction included based on gap filling. Small boxes list metabolites that can be taken up (green boxes) or secreted (brown boxes) under physiological conditions.
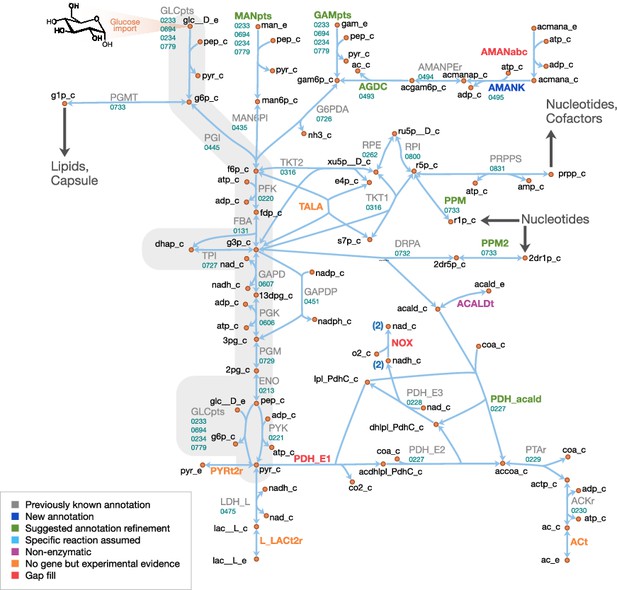
Central metabolism in JCVI-syn3A.
Map components and labels as in Figure 5. Big arrows denote incoming or outgoing connections to other parts of the metabolic network. For context, the node representing glucose transport has been labeled explicitly and glycolysis has been highlighted in gray.
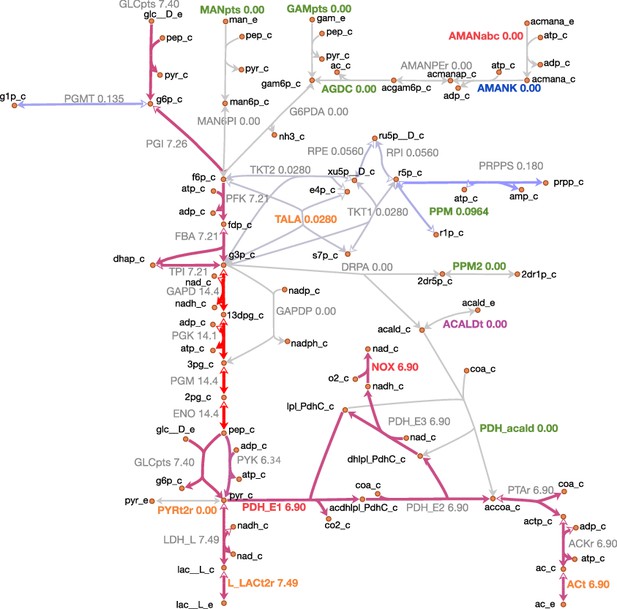
Steady-state fluxes through central metabolism in JCVI-syn3A.
Map components and labels as in Figure 5, with gene loci/gene-protein-reaction rules omitted. Numbers after reaction labels denote steady-state reaction fluxes in mmol gDW−1 h-1; edge color corresponds to the absolute value of the carried flux—gray to blue to purple to red, from low to high flux. For reversible reactions, the reaction progresses from the white to the filled arrowhead.
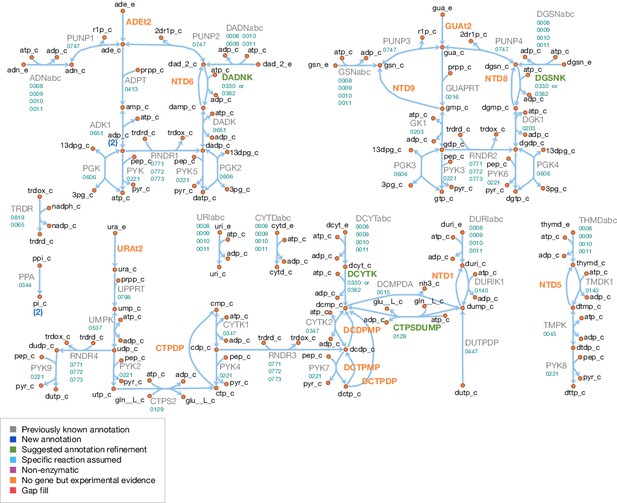
Nucleotide metabolism in JCVI-syn3A.
Map components and labels as in Figure 5.

Apparent dead-end of dUMP/deoxyuridine and possible solutions.
Internal metabolites are highlighted with cyan boxes, external ones with red boxes. Blue arrows denote reactions incorporated during model reconstruction—no reaction leads away from the dUMP/deoxyuridine pair. Red arrows denote hypothetical reactions that could possibly solve this dead-end. In the model, we have adopted the hypothetical CTP synthase reaction converting dUMP to dCMP (see also Figure 7; CTPSDUMP).
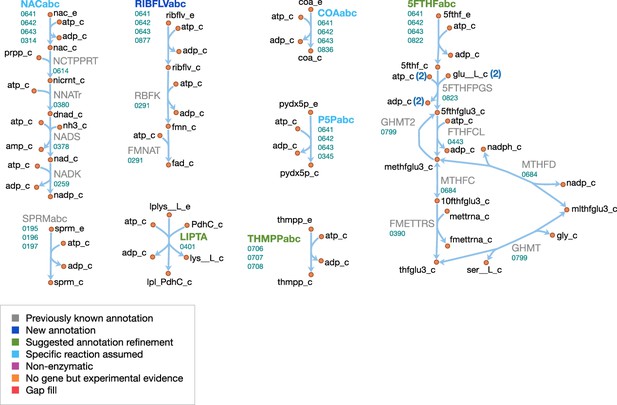
Cofactor metabolism in JCVI-syn3A.
Map components and labels as in Figure 5.
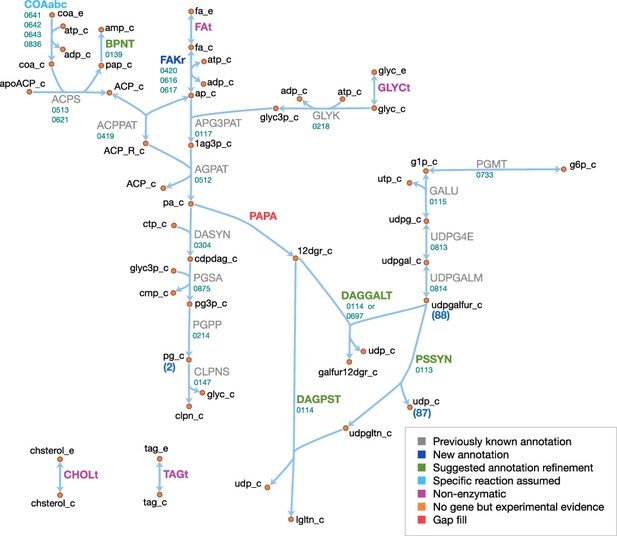
Lipid and capsule metabolism in JCVI-syn3A.
Map components and labels as in Figure 5.

Macromolecule metabolism in JCVI-syn3A.
Map components and labels as in Figure 5. The detailed (amino acid-specific) stoichiometry of the protein synthesis and degradation reactions can be found in Supplementary file 4. Protein synthesis reactions for the proteins explicitly included in the model (apo-ACP, dUTPase and PdhC) are analogous to the translation reaction shown and are therefore not included in the map.
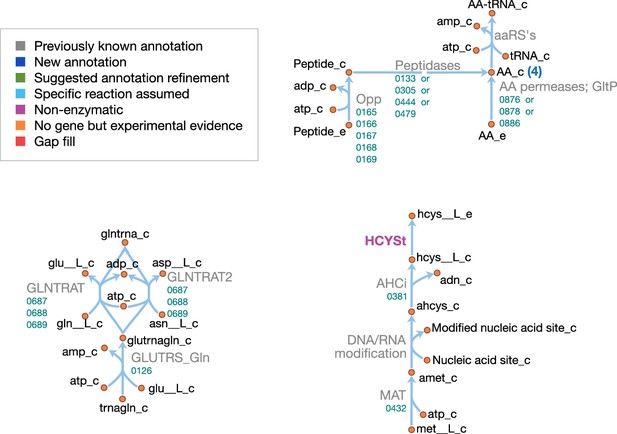
Amino acid metabolism in JCVI-syn3A.
Map components and labels as in Figure 5. As amino acid metabolism in JCVI-syn3A constitutes sets of analogous reactions (for each amino acid or peptide), we use generic reactions in the upper right part of the map. The ABC importer Opp catalyzes tetrapeptide uptake reactions in the model ([amino acid]4abc in Supplementary file 4); the AA permeases (incl. GltP) catalyze amino acid proton symport reactions ([amino acid]t2[p]r in Supplementary file 4). The peptidases catalyze peptide hydrolysis reactions ([amino acid]4P in Supplementary file 4). The aminoacyl tRNA synthetases (‘aaRS’s’ in the map) catalyze charging of tRNAs ([amino acid]TRS in Supplementary file 4). Synthesis of Gln-tRNAGln requires transamidation of initially mischarged Glu-tRNA and the corresponding reactions are shown on the lower left. In the -adenosylmethionine pathway on the lower right, we note that nucleic acid modification reactions (indicated by the edge labeled ‘DNA/RNA modification’) were not included in the model due to lack of sufficient information on kind and abundance of nucleic acid modifications in JCVI-syn3A.
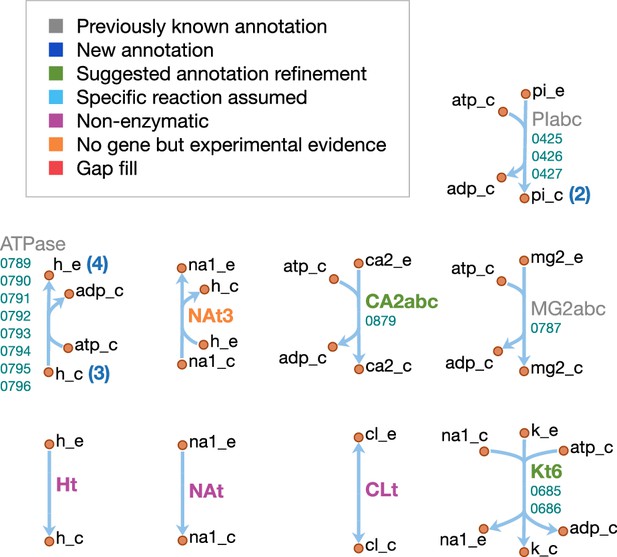
Ion transport reactions in JCVI-syn3A.
Map components and labels as in Figure 5.
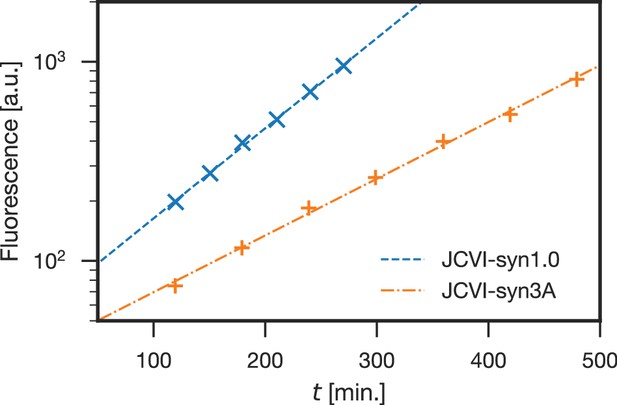
Comparison of growth curves of JCVI-syn1.0 and JCVI-syn3A.
JCVI-syn1.0 has a doubling time of 66 min (blue; ‘×' markers), whereas JCVI-syn3A has a doubling time of 105 min (orange; ‘+' markers). Doubling times () were calculated as described in Section 'Materials and methods', plotting fluorescence staining of cellular DNA vs. time, fitted by exponential regression curves. The regression curves for JCVI-syn1.0 and JCVI-syn3A have values of 0.9986 and 0.9976, respectively.
Comparison of FBA steady-state fluxes to maximal fluxes obtained from protein abundances and turnover numbers from BRENDA and the literature.
Map components and labels as in Figure 5, with reaction highlighting and gene loci/gene-protein-reaction rules omitted. Each edge is colored according to the ratio between and : Blue indicates , red indicates and green indicates that no could be obtained (because of either missing turnover number or missing protein abundance; or because reaction is not enzymatic to begin with).
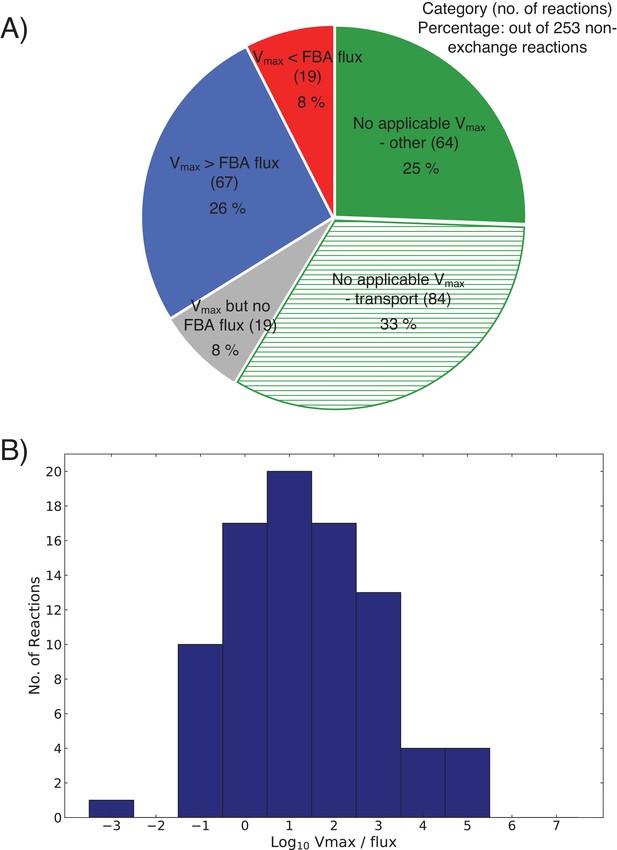
Statistics of FBA steady-state fluxes vs. maximal fluxes comparison (see Figure 15).
(A) Summary of vs. comparison over all 253 non-exchange reactions in the model. Red, blue, green: Meaning as in Figure 15. Green-striped: Subset of green set—reactions without that pertain to transport, which usually do not have an EC number associated with them. Gray: Reactions with in the FBA solution (thus always fulfilled). B: Histogram of over the blue and red subset in panel A.
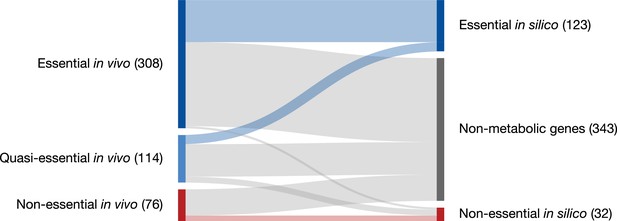
Partitioning of genes classified as essential, quasi-essential, and non-essential by transposon mutagenesis experiments into those which are in silico essential, in silico non-essential, and not modeled (‘Non-metabolic’).
All genes are included (i.e. also RNA genes and pseudogenes).
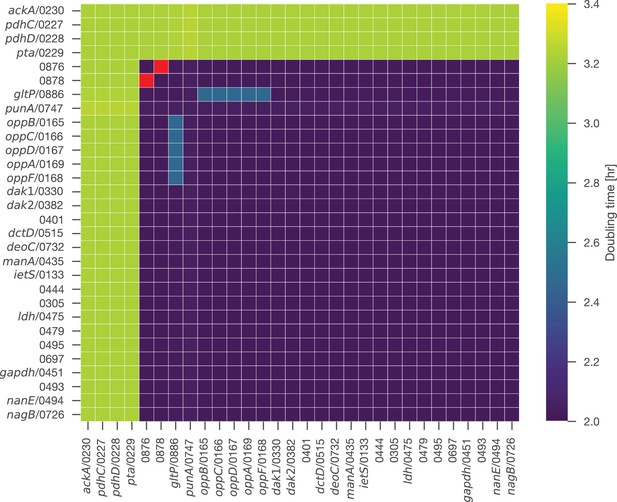
In silico double-gene knockouts between genes that are non-essential in single-gene knockouts.
Among the individually non-essential genes, a double knockout of the gene pair (0876, 0878) is the only lethal combination (red). This knockout corresponds to simultaneously removing both amino acid permeases, thus preventing cysteine uptake. Simultaneous knockout of the glutamate/aspartate permease gltP/0886 and any Opp gene (oppB/0165 through oppA/0169) is non-lethal in silico, as the model will under these circumstances produce glutamate through the hypothesized dUMP breakdown reaction CTPSDUMP and, to a lesser extent, through the reaction CTPS2 (both catalyzed by pyrG/0129). Glutamate production through pyrG/0129 is not expected to be able to meet cellular demands in vivo. If flux through CTPSDUMP is set to zero in the model, a double knockout of gltP/0886 and Opp becomes lethal in silico.
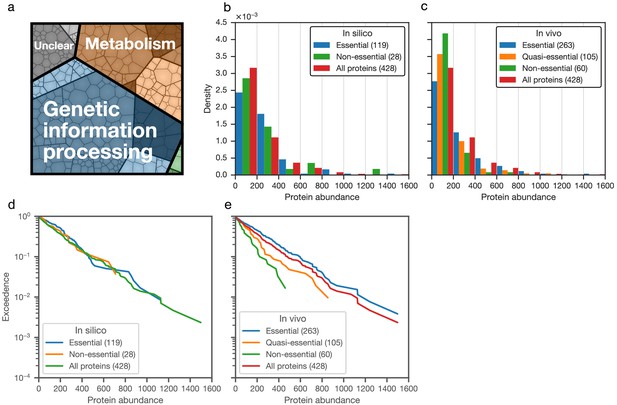
Distributions of absolute protein abundances (number of molecules per average cell) in JCVI-syn3A.
(a) Breakdown of the JCVI-syn3A proteome into functional classes. The area of each cell is proportional to its relative abundance. (b,c) Histograms of absolute protein abundances. (b) Absolute abundances of model-included metabolic proteins essential or non-essential in silico compared to all protein abundances. ‘Technical non-essential’ proteins are not included (see Section 'In silico gene knockouts and mapping to in vivo essentiality'). (c) Absolute abundances for proteins classified by in vivo essentiality from transposon mutagenesis experiments. (d,e) Exceedence plots of absolute abundances for proteins classified by in silico or in vivo essentiality. The exceedence at a given protein abundance value x is the fraction of the protein set displaying an abundance higher than x. (d) Model-included proteins (classified by in silico essentiality) compared to all proteins. (e) Proteins classified by in vivo (transposon-based) essentiality.
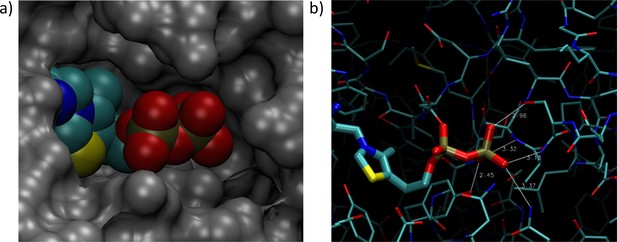
Thiamin diphosphate (ThDP) from the Mycoplasma hyorhinis Cypl crystal structure (pdb: 3EKI) overlaid onto the crystal structure of MG289 (pdb: 3MYU).
(Structures aligned using STAMP (Russell and Barton, 1992) in VMD (Humphrey et al., 1996; Eargle et al., 2006).) a): Space-filling view, with MG289 in gray and ThDP in color. The pyrophosphate tail of ThDP from the Cypl structure would have an appropriate cavity in MG289 as well. b): Visualization of hydrogen bonds for the same alignment. All possible hydrogen bonds are shown between potential donor and acceptor heavy atoms within 3.5 Å or less of each other. Even in the absence of the residues involved in pyrophosphate binding in Cypl (Sippel et al., 2009), the alignment suggests other side group and backbone interactions could still allow for pyrophosphate binding.
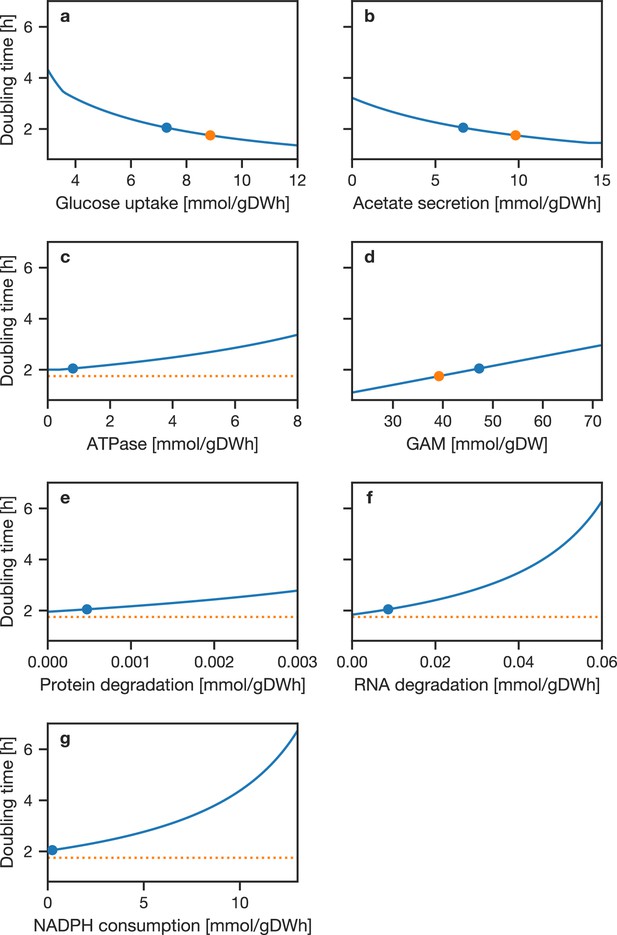
Sensitivity analysis of model doubling time with respect to model constraints.
In each panel, the stated parameter was varied over the indicated range and the model doubling time calculated while keeping all other constraints constant. (A:) Maximal glucose uptake. (B:) Maximal acetate secretion. (C:) ATPase ATP cost. D: GAM ATP cost. (E:) Protein degradation rate. (F:) RNA degradation rate. (G:) Imposed NADPH consumption. The blue circle marks the value used in the FBA model and resulting doubling time; the orange circle indicates the parameter that would yield the experimental doubling time. If there is no value of the parameter which would yield the experimental doubling time, a horizontal line is plotted.
Tables
Breakdown of protein coding genes in JCVI-syn3A into functional classes.
https://doi.org/10.7554/eLife.36842.009| Protein | Genes | Essentiality | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Functional hierarchy | % | # unique | % | # unique | # E | # Q | # N | # model | |
| Cellular processes | Cell Growth | 1.02 | 4 | 0.88 | 4 | 1 | 0 | 3 | 0 |
| Defense | 0.23 | 2 | 0.44 | 2 | 1 | 0 | 1 | 1 | |
| Subtotal | 1.25 | 6 | 1.33 | 6 | 2 | 0 | 4 | 1 | |
| Genetic information processing | DNA Maintenance | 5.07 | 38 | 8.41 | 38 | 25 | 9 | 4 | 3 |
| Folding, Sorting and Degradation | 9.58 | 25 | 5.53 | 25 | 18 | 7 | 0 | 7 | |
| Transcription | 3.92 | 14 | 3.32 | 15 | 8 | 5 | 2 | 0 | |
| Translation | 39.5 | 129 | 29.7 | 134 | 95 | 28 | 11 | 25 | |
| Subtotal | 58.1 | 206 | 46.9 | 212 | 146 | 49 | 17 | 35 | |
| Metabolism | Biosynthesis | 4.27 | 29 | 6.86 | 31 | 26 | 4 | 1 | 27 |
| Central Carbon Metabolism | 16.4 | 46 | 10.4 | 47 | 26 | 10 | 11 | 44 | |
| Energy Metabolism | 0.47 | 4 | 0.88 | 4 | 2 | 1 | 1 | 1 | |
| Membrane Transport | 9.37 | 54 | 12.6 | 57 | 37 | 16 | 4 | 46 | |
| Other Enzymes | 1.12 | 4 | 0.88 | 4 | 2 | 1 | 1 | 1 | |
| Subtotal | 31.6 | 137 | 31.6 | 143 | 93 | 32 | 18 | 119 | |
| Unclear | Kegg ortholog defined | 1.04 | 8 | 1.77 | 8 | 3 | 2 | 3 | 0 |
| No Kegg ortholog | 7.98 | 71 | 18.4 | 83 | 27 | 30 | 26 | 0 | |
| Subtotal | 9.02 | 79 | 20.1 | 91 | 30 | 32 | 29 | 0 | |
| Total | 100. | 428 | 100. | 452 | 271 | 113 | 68 | 155 | |
Genes modeled in the metabolic reconstruction.
The ‘MMSYN1_’ prefix on the locus tags has been omitted for brevity. The reaction column provides the specific reaction name or general description of the gene (if involved in multiple reactions). Reaction names may appear multiple times if there are multiple gene products that can catalyze that reaction. Column EssTn5 contains a if the gene is non-essential, a if it is quasi-essential, or a ■ if it has been determined to be essential through the transposon mutagenesis experiments. A dagger in this column indicates that the automatic essentiality assignment required manual intervention. Column EssFBA contains a if the gene is non-essential or a ■ if it has been determined to be essential through FBA. Loci marked with an asterisk are genes that are non-essential only ‘technically’ with respect to FBA (see Section 'In silico gene knockouts and mapping to in vivo essentiality'). The doubling times predicted by FBA for non-essential genes were all 2.02 hr, with the exception of single knockouts of loci pdhC/0227 through ackA/0230, which all had doubling times of 3.22 hr; locus punA/0747 with a doubling time of 2.04 hr; and locus gltP/0886 with a doubling time of 2.03 hr.
| Locus | Reaction | EssTn5 | EssFBA | Locus | Reaction | EssTn5 | EssFBA | Locus | Reaction | EssTn5 | EssFBA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid metabolism | Cofactor metabolism | 0798 | UPPRT | ■ | ■ | ||||||
| 0381* | AHCi | 0823 | 5FTHFPGS | ■ | 0330 | dAdn kinase 1 | |||||
| 0163 | ALATRS | ■ | ■ | 0390 | FMETTRS | ■ | 0382 | dAdn kinase 2 | |||
| 0535 | ARGTRS | ■ | ■ | 0291 | FMNAT | ■ | ■ | Transport | |||
| 0076 | ASNTRS | ■ | ■ | 0443 | FTHFCL | ■ | ■ | 0822 | 5FTHFabc | ■ | |
| 0287 | ASPTRS | ■ | ■ | 0799 | GHMT | ■ | 0876 | AA permease 1 | |||
| 0837 | CYSTRS | ■ | ■ | 0684 | MTHFC | ■ | 0878 | AA permease 2 | |||
| 0687 | GLNTRAT | ■ | ■ | 0259 | NADK | ■ | ■ | 0789 | ATPase | ■† | ■ |
| 0688 | GLNTRAT | ■ | ■ | 0378 | NADS | ■ | ■ | 0790 | ATPase | ■ | ■ |
| 0689 | GLNTRAT | ■ | ■ | 0614 | NCTPPRT | ■ | ■ | 0791 | ATPase | ■ | ■ |
| 0126 | GLUTRS_Gln | ■ | ■ | 0380 | NNATr | ■ | ■ | 0792 | ATPase | ■ | ■ |
| 0405 | GLYTRS | ■ | ■ | Lipid metabolism | 0793 | ATPase | ■ | ■ | |||
| 0288 | HISTRS | ■ | ■ | 0621 | ACP | ■ | ■ | 0794 | ATPase | ■ | ■ |
| 0519 | ILETRS | ■ | ■ | 0419 | ACPPAT | ■ | ■ | 0795 | ATPase | ■ | ■ |
| 0634 | LEUTRS | ■ | ■ | 0513 | ACPS | ■ | ■ | 0796 | ATPase | ■ | ■ |
| 0064 | LYSTRS | ■ | ■ | 0512 | AGPAT | ■ | ■ | 0879 | CA2abc | ■ | ■ |
| 0432 | MAT | ■ | 0117 | APG3PAT | ■ | ■ | 0836 | COAabc | ■ | ■ | |
| 0012 | METTRS | ■ | ■ | 0139 | BPNT | ■ | 0642 | EcfA | ■ | ■ | |
| 0528 | PHETRS | ■ | ■ | 0147 | CLPNS | ■ | ■ | 0643 | EcfA | ■ | ■ |
| 0529 | PHETRS | ■ | ■ | 0697 | DAGGALT | 0641 | EcfT | ■ | ■ | ||
| 0282 | PROTRS | ■ | ■ | 0114 | DAGPST/DAGGALT | ■ | ■ | 0233 | GLCpts | ■ | ■ |
| 0133 | Peptidase 1 | 0304 | DASYN | ■ | ■ | 0234 | GLCpts | ■ | ■ | ||
| 0305 | Peptidase 2 | 0420 | FAKr | ■ | ■ | 0694 | GLCpts | ■ | ■ | ||
| 0444 | Peptidase 3 | 0616 | FAKr | ■ | ■ | 0779 | GLCpts | ■ | ■ | ||
| 0479 | Peptidase 4 | 0617 | FAKr | ■ | 0886 | GltP | |||||
| 0061 | SERTRS | ■ | ■ | 0115 | GALU | ■ | ■ | 0685 | Kt6 | ■ | ■ |
| 0222 | THRTRS | ■ | ■ | 0218 | GLYK | ■ | ■ | 0686 | Kt6 | ■ | ■ |
| 0308 | TRPTRS | ■ | ■ | 0733 | PGMT/PPM | ■ | ■ | 0401 | LIPTA | ||
| 0613 | TYRTRS | ■ | ■ | 0214 | PGPP | ■ | 0787 | MG2abc | ■ | ■ | |
| 0260 | VALTRS | ■ | ■ | 0875 | PGSA | ■ | ■ | 0314 | NACabc | ■ | ■ |
| Central metabolism | 0113 | PSSYN | ■ | ■ | 0165 | Opp | |||||
| 0230 | ACKr | ■ | 0813 | UDPG4E | ■ | ■ | 0166 | Opp | |||
| 0493 | AGDC | 0814 | UDPGALM | ■ | ■ | 0167 | Opp | ||||
| 0495 | AMANK | Macromolecules | 0168 | Opp | |||||||
| 0494 | AMANPEr | 0394 | Lon | ■ | 0169 | Opp | |||||
| 0732 | DRPA | 0650 | Met peptidase | ■ | ■ | 0345 | P5Pabc | ■ | ■ | ||
| 0213 | ENO | ■ | ■ | 0201 | Pept. deformylase | ■ | ■ | 0425 | PIabc | ■ | ■ |
| 0131 | FBA | ■ | ■ | Nucleotide metabolism | 0426 | PIabc | ■ | ■ | |||
| 0726 | G6PDA | 0651 | (D)ADK | ■ | ■ | 0427 | PIabc | ■ | ■ | ||
| 0607 | GAPD | ■ | ■ | 0413 | ADPT | ■ | ■ | 0877 | RIBFLVabc | ■ | ■ |
| 0451 | GAPDP | ■ | 0129 | CTPS | ■ | ■ | 0008 | RNS | ■ | ■ | |
| 0475 | LDH_L | ■ | 0347 | CYTK | ■ | 0009 | RNS | ■ | ■ | ||
| 0435 | MAN6PI | 0515 | DCMPDA | 0010 | RNS | ■ | ■ | ||||
| 0227 | PDH_E2/_acald | 0447 | DUTPDP | ■ | ■ | 0011 | RNS | ■ | ■ | ||
| 0228 | PDH_E3 | 0203 | GK | ■ | ■ | 0195 | SPRMabc | ■ | |||
| 0220 | PFK | ■ | ■ | 0216 | GUAPRT | ■ | 0196 | SPRMabc | ■ | ||
| 0445 | PGI | ■ | ■ | 0747 | PNP | 0197 | SPRMabc | ■ | |||
| 0606 | PGK | ■ | ■ | 0344 | PPA | ■ | ■ | 0706 | THMPPabc | ■ | |
| 0729 | PGM | ■ | ■ | 0771 | RNDR | ■ | 0707 | THMPPabc | ■ | ||
| 0831 | PRPPS | ■ | ■ | 0772 | RNDR | ■ | ■ | 0708 | THMPPabc | ■ | |
| 0229 | PTAr | ■ | 0773 | RNDR | ■ | ||||||
| 0221 | PYK | ■ | ■ | 0140 | TMDK1/DURIK1 | ■ | ■ | ||||
| 0262 | RPE | ■ | 0045 | TMPK | ■ | ■ | |||||
| 0800 | RPI | ■ | 0065 | TRDR | ■ | ■ | |||||
| 0316 | TKT | ■ | 0819 | TRDR | ■ | ■ | |||||
| 0727 | TPI | ■ | ■ | 0537 | UMPK | ■ | ■ | ||||
Cellular ATP expenses by category (in percent of total ATP consumption).
https://doi.org/10.7554/eLife.36842.025| Category | ATP expense [%] |
|---|---|
| Nucleotide metabolism | 3.6 |
| Pentose phosphate pathway | 1.7 |
| Lipid metabolism | 0.7 |
| Cofactor metabolism | 0.1 |
| Transport | 3.4 |
| GAMMacromolecules | 18 |
| GAMtRNA charging | 16 |
| GAMNonquant | 41 |
| NGAMTurnover | 13 |
| NGAMATPase | 2.7 |
Confusion matrices for gene essentiality prediction.
‘All genes’ denotes agreement/disagreement between the model prediction and the transposon mutagenesis experiment considering all genes in the metabolic reconstruction (excluding the two ‘technical non-essential’ genes). ‘Excluding AA’ repeats the same comparison as ‘All genes’, with genes for amino acid utilization (uptake and peptidase genes) excluded. See Table 2 for individual gene essentialities in silico and in vivo.
| All genes | Excluding AA | |||
|---|---|---|---|---|
| Exp. / Model | Essential | Non-essential | Essential | Non-essential |
| Essential | 101 | 4 | 101 | 4 |
| Quasi-essential | 22 | 14 | 22 | 4 |
| Non-essential | 0 | 12 | 0 | 10 |
Accuracy, sensitivity, specificity and Matthews correlation coefficient calculated for several scenarios.
QE as E: Treating in vivo quasi-essentials as essentials; QE as NE: Treating quasi-essentials as non-essentials; No QE genes: Excluding all genes quasi-essential in vivo; QE as E, no AA genes: Excluding genes related to amino acid utilization, and treating quasi-essentials as essentials; QE as NE, no AA genes: Excluding genes related to amino acid utilization, and treating quasi-essentials as non-essentials.
| Accuracy | Sensitivity | Specificity | MCC | |
|---|---|---|---|---|
| QE as E | 88% | 87% | 100% | 0.59 |
| QE as NE | 83% | 96% | 54% | 0.59 |
| No QE genes | 97% | 96% | 100% | 0.85 |
| QE as E, no AA gene | 94% | 94% | 100% | 0.72 |
| QE as NE, no AA genes | 82% | 96% | 39% | 0.46 |
Summary of features of the metabolic model for JCVI-syn3A.
‘Non-pseudo’ reactions exclude exchange, biomass and macromolecule reactions. ‘Annotation-supported’ includes all non-pseudo reactions that have a gene assigned. ‘Passive’ reactions are transport processes assumed to take place without protein mediation. The meaning of ‘technical’ non-essential genes is explained in Section 'In silico gene knockouts and mapping to in vivo essentiality'.
| Model overview | Genes | 155 | |
| Genome coverage | 31% | ||
| Metabolites | 304 | ||
| Reactions (total) | 338 | ||
| Reactions (non-pseudo) | 244 | ||
| Reaction breakdown (% of non-pseudo) | Annotation-supported | 209 | 86% |
| Passive | 14 | 6% | |
| Gap fills with exp. evidence | 17 | 7% | |
| Gap fills without exp. evidence | 4 | 2% | |
| Supported (annotation/exp./passive) | 240 | 98% | |
| Essentiality insilico | Essential genes | 123 | 79% |
| Non-essential genes | 30 | 19% | |
| ‘Technical' non-essential genes | 2 | 1% | |
| Essentiality invivo | Essential genes | 308 | 62% |
| Quasi-essential genes | 114 | 23% | |
| Non-essential genes | 76 | 15% |
List of suggested gene removal experiments.
https://doi.org/10.7554/eLife.36842.032| Gene(s) | Description | Remark |
|---|---|---|
| manA/0435 | Mannose 6-phosphate isomerase | |
| deoC/0732 | Deoxyribose phosphate aldolase | |
| nanE/0494, 0495, nagB/0726 | N-acetylmannosamine 6-phosphate branch | |
| pdhC/0227, pdhD/0228, 0401 | pdhCD and proposed lipoate importer | |
| dak1/0330, dak2/0382 | Deoxynucleoside kinases | Synthetic lethality possible |
| ietS/0133 | Peptidase | |
| 0876 | Amino acid permease | |
| folT in JCVI-syn3A | Folate uptake and usage | Competition experiment with wild type to probe fitness cost in JCVI-syn3A |
| folT in JCVI-syn1.0 | Folate uptake and usage | Competition experiment with wild type to probe fitness cost in JCVI-syn1.0 |
List of suggested experiments, with rationale behind each suggestion.
https://doi.org/10.7554/eLife.36842.033| Experiment | Rationale |
|---|---|
| Nutrient utilization | |
| Detect lipoylpeptide uptake | Lipoate is cofactor for PDH, whose functionality is unclear after E1 subunit deletion. |
| Detect nucleotide uptake | Activity reported for M. mycoides capri without gene assignment; alternative routes present in JCVI-syn3A, but activity not ruled out. |
| Detect nucleobase uptake | Activity reported for M. mycoides capri, and uracil uptake essential in model. |
| Demonstrate growth on thiamine diphosphate | Structural data and deletion of thiamine diphosphokinase suggest thiamine diphosphate (ThDP) uptake. Inability to grow on ThDP would imply unidentified kinase activity. |
| Demonstrate growth on pyridoxal phosphate | With no pyridoxal kinase identified, growth on pyridoxal phosphate (P5P) assumed; inability to grow on P5P would imply unidentified kinase activity. |
| Demonstrate growth on acetylmannosamine | Reconstruction suggests operon 0493 through 0495 to be nagA/nanE/nagC; growth on acetylmannosamine would support assignment and imply uptake capability. |
| Demonstrate growth on mannose or glucosamine | Literature suggests mannose and glucosamine import through PtsG; and downstream enzymes are present. |
| Metabolite production/secretion | |
| Detect production of acetate | Acetate pathway has been partially removed, but several of the remaining enzymes remain essential in transposon mutagenesis experiments. |
| Investigate production of lipogalactan capsule | Genetic evidence suggests capsule is still being produced. |
| Detect secretion of deoxyuridine or dUMP | Deoxyuridine/dUMP is currently dead-end; secretion would be one possible solution. |
| Enzymatic activity | |
| Detect pyruvate oxidation | Conversion of pyruvate to acetyl-CoA by truncated PDH complex has been assumed for the time being but is not supported by experiment. |
| Detect oxidation of acetaldehyde | Conversion of acetaldehyde to acetyl-CoA would provide alternative explanation for presence of truncated PDH complex in JCVI-syn3A in spite of deletion of first subunit. |
| Detect NOX activity | NADH oxidase (NOX) has been deleted but activity would be necessary for PDH activity against both pyruvate and acetaldehyde. |
| Detect transaldolase activity | Activity is essential in model and has been detected in M. mycoides capri; no known gene in any mycoplasma though. |
| Detect sedoheptulose-1,7-bisphosphate phosphatase activity | Reaction would provide bypass to transaldolase reaction. |
| Detect phosphatidate phosphatase activity | Gene present in M. pneumoniae and reaction is missing link to diacylglycerol, but no gene identified in JCVI-syn3A. |
| Assess phosphatase activity against dCTP, dCDP, GMP, dAMP, dGMP, dUMP, dTMP; pyrophosphatase activity against CTP, dCTP | Activities observed in M. mycoides capri but no gene identified in JCVI-syn3A. |
| Detect deoxyuridine phosphorylase activity | Gene has been removed in JCVI-syn3A (MMSYN1_0734), but deoxyuridine/dUMP is currently dead-end, raising the question whether function is carried out by some other gene. |
| Detect thymidylate synthase activity | Extremely low activity detected in M. mycoides mycoides SC, but no gene identified. Reaction would be alternative solution to deoxyuridine/dUMP dead-end. |
| Specific gene function | |
| Determine substrates for deoxynucleoside kinase dak2/0382 | Presence of dak2/0382 in addition to the characterized dak1/0330 suggests different substrate profile. |
| Verify CTPS activity against dUMP | CTPS (pyrG/0129) converting dUMP to dCMP seems most plausible solution to deoxyuridine/dUMP dead-end. |
| Check PGPP activity against phosphatidate | Activity observed for PgpB in E. coli; activity for PGPP (pgpA/0214) would provide missing link to diacylglycerol in apparent absence of phosphatidate phosphatase gene. |
| Check UMPK activity against CMP and dCMP | Substrate profile for UMP kinase similar to eukaryotic enzyme could explain quasi-essentiality of CMP kinase. |
| Change of expression levels | |
| Knock out deoxynucleoside kinases and over express RNDR | RNDR and deoxynucleoside kinases provide redundant routes to deoxydinucleotides, suggesting one pathway might be sufficient if expression increased. |
| Reduce expression of RNDR and knock out putative dUTPase simultaneously | RNDR is currently only known source of dUTP. If RNDR knockdown would make the putative dUTPase gene dut/0447 nonessential as well, this would corroborate the putative assignment. |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | JCVI-syn3A | JCVI; this article | GenBank accession number:CP016816.2 | [1] |
| Strain, strain background | JCVI-syn1.0 | doi:10.1126/science.1190719 | GenBank accession number:CP002027.1 | [1] |
| Genetic reagent | terTufPuro transposome | doi:10.1126/science.aad6253 | Constructed by the JCVI | |
| Genetic reagent | Yeast tRNA | Life Technologies, Carlsbad, CA, USA | 15-401-029 | |
| Genetic reagent | EZ-Tn5-Transposase | Lucigen, Madison, WI, USA | TNP92110 | |
| Sequence-based reagent | Custom forward primer | Integrated DNA technologies, San Diego, CA, USA | [2] | |
| Commercial assay or kit | Nextera XT DNA library preparation kit | Illumina, San Diego, CA, USA | FC-131-1024 | |
Chemical compound, drug | Quant-iT PicoGreen | Molecular Probes, Eugene, OR, USA | P7589 | |
| Chemical compound, drug | Puromycin | Molecular Probes, Eugene, OR, USA | A1113802 | |
| Software, algorithm | COBRApy | doi:10.1186/1752-0509-7-74 | ||
| Software, algorithm | CLC Genomics Workbench | QIAGEN Bioinformatics, Redwood City, CA, USA | ||
| Software, algorithm | ProteomeDiscoverer 2.1.0.81 | Thermo Fisher Scientific |
-
[1] Bacterial strains JCVI-syn3A and JCVI-syn1.0 will be made available to qualified researchers by the JCVI and Synthetic Genomics, Inc under a material transfer agreement. Note that United States scientists must obtain a United States Veterinary Permit for Importation and Transportation of Controlled Materials and Organisms and Vectors from the U.S. Department of Agriculture Animal and Plant Health Inspection Service. The organisms require Biosafety Level 2 containment.
[2] Used for marker-specific sequencing with PCR; sequence under ‘Tn5 mutagenesis–Experimental method'.
Reconstructed biomass composition of JCVI-syn3A, listing the fraction of each component as percent of cellular dry mass.
https://doi.org/10.7554/eLife.36842.047| Category | Component | Fraction [%] |
| Macromolecules | Protein | 54.727 |
| Total: 76.521 % | RNA | 16.274 |
| DNA | 5.5 | |
| Acyl carrier protein | 0.018 | |
| dUTPase | 0.003 | |
| Lipids & capsule | Lipogalactan capsule | 6.368 |
| Total: 17.563 % | Phosphatidylglycerol | 2.944 |
| Cardiolipin | 2.944 | |
| Cholesterol | 1.534 | |
| Diacylglycerol | 1.366 | |
| Gal-DAG | 1.31 | |
| Triacylglycerol | 0.549 | |
| Fatty acid | 0.549 | |
| Small molecules & ions | Potassium | 3.285 |
| Total: 5.916 % | Phosphate | 0.375 |
| Chloride | 0.198 | |
| ATP | 0.167 | |
| L-lysine | 0.133 | |
| Sodium | 0.131 | |
| Spermine | 0.128 | |
| L-isoleucine | 0.115 | |
| GTP | 0.115 | |
| L-leucine | 0.112 | |
| UTP | 0.106 | |
| L-glutamate | 0.085 | |
| L-valine | 0.083 | |
| L-asparagine | 0.083 | |
| Small molecules | L-alanine | 0.077 |
| & ions (cont’d) | L-aspartate | 0.072 |
| L-threonine | 0.071 | |
| L-serine | 0.071 | |
| Glycine | 0.068 | |
| CTP | 0.053 | |
| L-phenylalanine | 0.049 | |
| L-glutamine | 0.048 | |
| L-arginine | 0.043 | |
| L-tyrosine | 0.041 | |
| L-proline | 0.035 | |
| L-methionine | 0.026 | |
| Magnesium | 0.019 | |
| L-histidine | 0.019 | |
| Calcium | 0.019 | |
| FAD | 0.016 | |
| 5,10-meTHF(Glu)3 | 0.015 | |
| CoA | 0.012 | |
| Thiamin diphosphate | 0.009 | |
| NADP+ | 0.008 | |
| L-tryptophan | 0.008 | |
| L-cysteine | 0.008 | |
| Pyridoxal phosphate | 0.005 | |
| dTTP | 0.003 | |
| dATP | 0.003 | |
| dCTP | 0.002 | |
| dGTP | 0.001 |
Additional files
-
Supplementary file 1
Gene classification hierarchy, model reaction breakdown and protein gene breakdowns for M. pneumoniae and E. coli.
- https://doi.org/10.7554/eLife.36842.034
-
Supplementary file 2
Transposon insertion nucleotide positions.
- https://doi.org/10.7554/eLife.36842.035
-
Supplementary file 3
Transposon insertion counts and assignment of gene essentiality from both transposon mutagenesis and FBA.
- https://doi.org/10.7554/eLife.36842.036
-
Supplementary file 4
Reactions and metabolites included in the metabolic reconstruction.
- https://doi.org/10.7554/eLife.36842.037
-
Supplementary file 5
Data from proteomics experiments.
- https://doi.org/10.7554/eLife.36842.038
-
Supplementary file 6
Comparison of the JCVI-syn3A metabolic reconstruction to that of M. pneumoniae published by Wodke et al. (2013).
- https://doi.org/10.7554/eLife.36842.039
-
Supplementary file 7
Flux constraints derived from proteomics and turnover numbers and comparison to FBA fluxes.
- https://doi.org/10.7554/eLife.36842.040
-
Supplementary file 8
Known metabolic reactions removed during genome minimization from JCVI-syn1.0 to JCVI-syn3A.
- https://doi.org/10.7554/eLife.36842.041
-
Supplementary file 9
FBA model in sbml format.
- https://doi.org/10.7554/eLife.36842.042
-
Supplementary file 10
FBA model in json format.
- https://doi.org/10.7554/eLife.36842.043
-
Supplementary file 11
ESCHER network map in json format.
- https://doi.org/10.7554/eLife.36842.044
-
Transparent reporting form
- https://doi.org/10.7554/eLife.36842.045





