A bacterial immunomodulatory protein with lipocalin-like domains facilitates host–bacteria mutualism in larval zebrafish
Figures
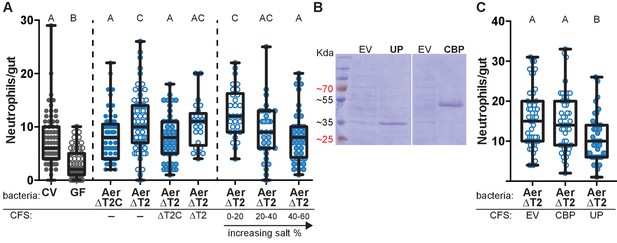
Aeromonas secretes a protein that regulates intestinal neutrophil response to a general model of inflammation.
(A) Intestinal neutrophil response to conventional (CV), germ free (GF), and mono-associations of wild-type Aeromonas (Aer ΔT2C), and an isogenic mutant of the type II secretion system (ΔT2). ΔT2 induces a greater neutrophil response that is rescued by addition of cell free supernatant (CFS) from the wild-type Aeromonas strain. Ammonium sulfate fractionation narrowed potential candidates down to two proteins, an unidentified protein (UP) or chitin binding protein (CBP). (B) E. coli BL21 carrying pET21b expression vector overexpression of the candidate proteins of interest. (C) Addition of CFS containing UP rescued the increased neutrophil response induced by ΔT2. EV: empty vector control CFS. Letters denote p<0.05, ANOVA followed by Tukey’s post hoc test. Each box represents the first to third quartiles, center bar the median, and whiskers the maximum and minimum. Each dot represents one fish; data collected from at least two independent experiments; n ≥ 24.
-
Figure 1—source data 1
Aeromonas secretes a protein that regulates intestinal neutrophil response to a general model of inflammation.
- https://doi.org/10.7554/eLife.37172.005
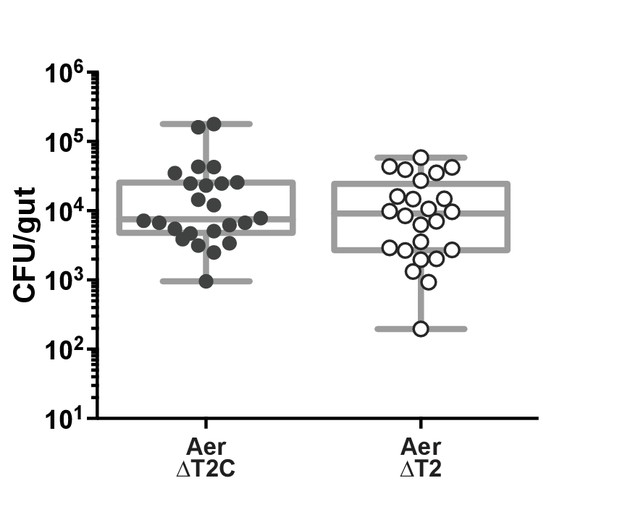
Colonization of A. veronii strain Hm21 type two secretion mutant.
(A) veronii strain Hm21 isolates ΔT2, the type two secretion mutant, and ΔT2C, the compliment isolate with a wild-type phenotype, colonize the fish to similar levels. Each dot represents one fish. n ≥ 24, from two independent experiments.
-
Figure 1—figure supplement 1—source data 1
Colonization of A. veronii strain Hm21 type 2 secretion mutant.
- https://doi.org/10.7554/eLife.37172.006
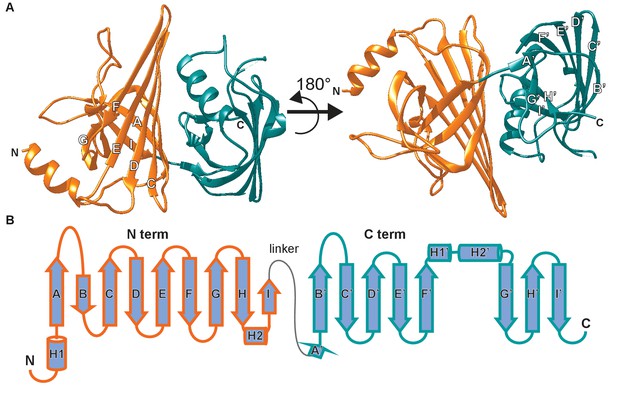
The crystal structure of AimA reveals two calycin domains.
(A) The 2.3 Å structure of AimA displays two calycin domains (PDB ID 6B7L). (B) The amino terminal domain (orange) of AimA is connected by a short linker to the carboxy terminal domain (teal) and both contain an eight-stranded full (C-term) or partial (N-term) β-barrel.

Structural homology of the two domains of AimA to avidins.
(A) Structural overlay of N-term domain of AimA with top structural homology hit from PDBeFold search (Supplemental Table 2), Streptomyces avindinii streptavidin (PDB ID 1KL3). (B) Structural overlay of C-term domain of AimA with top structural homology hit from PDBeFold search (Supplemental Table 2), Gallus gallus avidin (PDB ID 1LEL). (C) Schematic of the biotin binding colorimetric assay. Biotin has higher affinity for avidins than HABA does, so it replaces HABA in the binding site, thereby decreasing the absorbance at 500 nm. (D) Results from the biotin binding colorimetric assay. Biotin replaces HABA binding in avidin, corresponding to a decrease in absorbance when biotin is present. With AimA, there is no difference in absorbance with or without biotin, indicating no biotin binding.
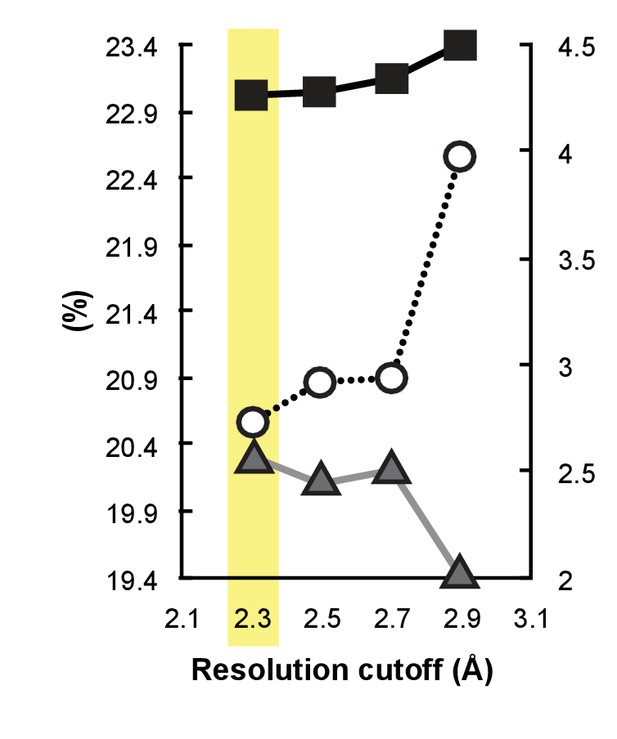
A paired refinement demonstrates that the extended resolution improves the quality of the model.
Shown are Rfree (black line, solid squares), Rwork, (gray line, solid triangles), and Rfree-Rwork (dotted line, open circles) calculated at 2.9 Å for paired refinements in which the model was first refined against either 2.9, 2.7, 2.5, or 2.3 Å data. The chosen resolution cutoff of 2.3 Å (highlighted in yellow) show a decrease in Rfree and increase in Rwork, which proves that using the extra resolution improves the model.
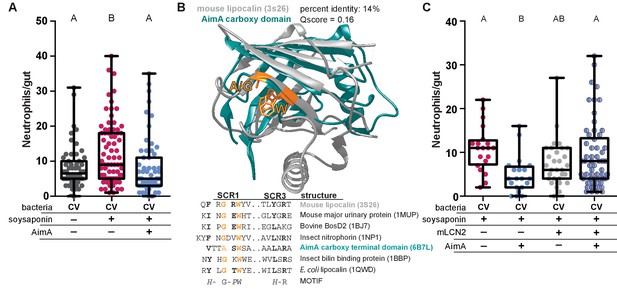
AimA contains regions structurally conserved with the lipocalin family and reduces soysaponin induced inflammation.
(A) Feeding zebrafish soy saponin induces increased intestinal neutrophil response, and treating those fish with 100 ng/mL purified AimA prevents the increased intestinal neutrophil response. Letters indicate significantly different groups; ANOVA with multiple comparisons. Each dot represents one fish; data collected from at least two independent experiments; n ≥ 24. (B) Structural overlay of mouse lipocalin (PDB ID 3S26) and AimA C-term domain using PDBeFold. Qscore is a structural overlay quality score that takes into account both the root mean standard deviation (RMSD) of the Cα carbons and the alignment length. Qscore of 1 is perfect alignment, 0 is no alignment. The residues highlighted in orange are conserved in both the sequences and the structures (see table below). Displayed are the sequences of Structurally Conserved Regions (SCR) 1 and 3 of a representative set of kernel and outlier lipocalins, with PDB IDs in parenthesis. The C-term domain of AimA is included and contains a subset of conserved SCR residues. Bold residues are conserved across the sequences. Orange residues are conserved across the sequences and in the structures of mLCN and the C-term domain of AimA. (C) Treatment of conventionally raised (CV) fish with soysaponin and lipocalin prevents AimA from reducing the neutrophil response. Each dot represents one fish; n ≥ 20 from at least three independent experiments. Letters indicate significantly different groups; ANOVA with multiple comparisons.
-
Figure 3—source data 1
AimA contains regions structurally conserved with the lipocalin family and reduces soysaponin induced inflammation.
- https://doi.org/10.7554/eLife.37172.014
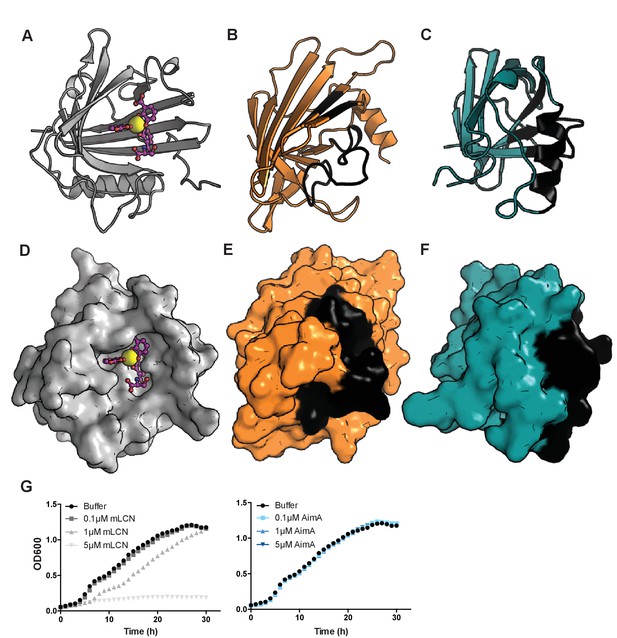
AimA does not bind enterobactin.
(A) Shown is a ribbon diagram of the structure of human LCN2 (PDB ID 1L6M) with ferric enterobactin bound. Enterobactin is shown in pink with oxygen in red, nitrogen in blue, and iron as a yellow ball. (B) After overlay with LCN2 from panel A, shown is a ribbon diagram of the structure of AimA N-term domain in the same orientation. The regions colored black indicate residues that would occlude enterobactin binding. (C) After overlay with LCN2 from panel A, shown is a ribbon diagram of the structure of AimA C-term domain in the same orientation. The regions colored black indicate residues that would occlude enterobactin binding. (D–F) Shown are the surface representations of LCN2, N-term AimA and C-term AimA from panels A-C, respectively. Note the deep calyx formed by LNC2 for enterobactin binding is missing in both the N-term and C-term AimA domains. (G) E. coli K12 grown in the presence of the iron chelator dipyridyl experiences a dose dependent growth inhibition upon addition of purified mouse LCN2 protein (left), but does not experience growth inhibition upon addition of the same range of purified AimA concentrations to the E. coli cultures (right).
-
Figure 3—figure supplement 1—source data 1
AimA does not bind enterobactin.
- https://doi.org/10.7554/eLife.37172.015
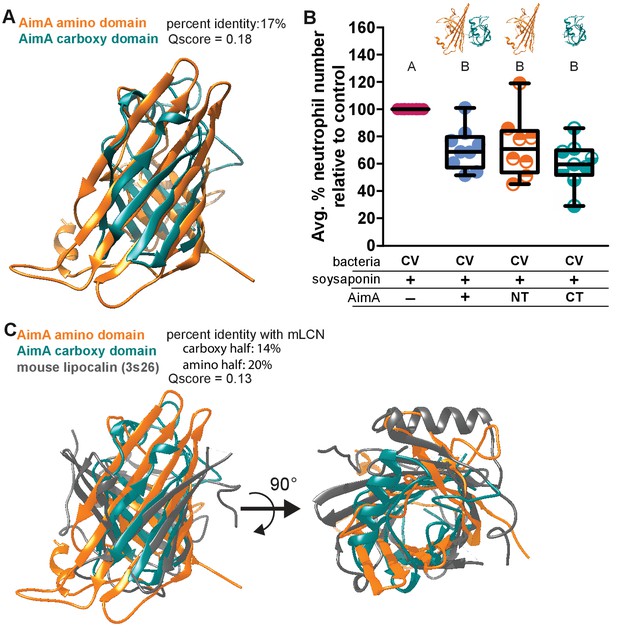
Both domains of AimA retain neutrophil modulating function.
(A) Overlay of C-term and N-term domains of AimA using PDBeFold. (B) Conventionally raised (CV) fish fed soysaponin (SS) and treated with either purified full-length AimA or purified N-term (NT) or C-term (CT). Each dot represents the average percent of neutrophil influx in a flask of 15 fish from the average neutrophil influx of a control flask (soysaponin only) of 15 fish. n ≥ 9 flasks from at least three independent experiments. Letters indicate significantly different groups; ANOVA with multiple comparisons. (C) Overlay of C- and N-term domains of AimA and mLCN using PDBeFold.
-
Figure 4—source data 1
Both domains of AimA retain neutrophil modulating function.
- https://doi.org/10.7554/eLife.37172.018
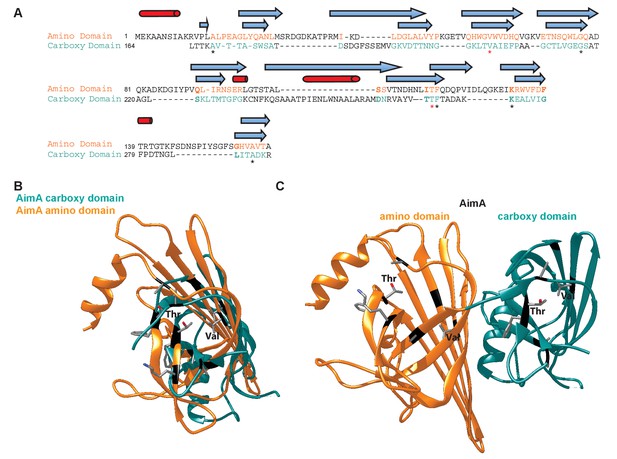
Structural comparison of N-term and C-term domains of AimA.
(A) Structure-based sequence alignment of N-term and C-term domains of AimA. Black text indicates regions that don’t align in the structures, colored text are regions that do. Blue arrows, β-strands, and red cylinders, α-helices, above the text represent the secondary structure, with the top symbols corresponding to the N-term domain. Stars show the seven residues that align in the structures, with red stars indicating the Val and Thr that may be functionally relevant. (B) Overlay of N-term (orange) and C-term (teal) domain of AimA. The seven structurally conserved residues are mapped on and shown as sticks (gray = carbon, red = oxygen, blue = nitrogen). (C) Full length AimA structure with the seven structurally conserved regions shown as sticks.
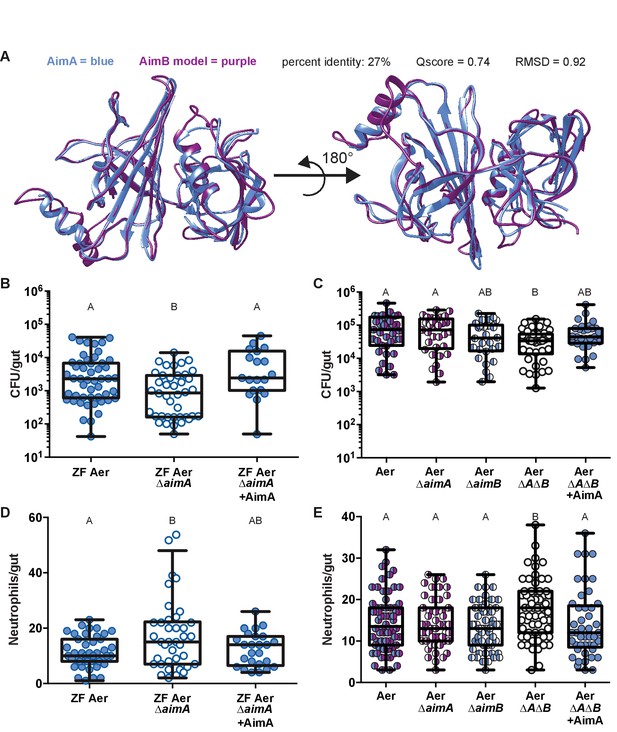
AimA reduces neutrophil influx and promotes colonization of Aeromonas.
(A) Structural overlay (PDBeFold) of AimA (blue) and the model of AimB (purple) generated by I-TASSER using AimA as a threading structure. (B) Colonization level of wild-type ZF Aeromonas and ZF Aer ΔAimA. The colonization defect of ZF Aer ΔAimA is rescued by treatment with 100 ng/mL purified AimA. (C) Colonization level of wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each of the single mutants colonizes as well as wild type, while the double mutant has a significantly reduced colonization level. This phenotype is rescued by treatment with 100 ng/mL purified AimA. (D) Intestinal neutrophil response to ZF Aer and the colonization defect of ZF Aer ΔAimA. (E) Intestinal neutrophil response to wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each of the single mutants induces a similar neutrophil response to wild type, while the double mutant induces significantly greater response. This phenotype is rescued by treatment with 100 ng/mL purified AimA for both Aeromonas isolates. For all graphs, each dot represents one fish; n ≥ 23 from at least three independent experiments. Letters indicate significantly different groups, ANOVA with multiple comparisons.
-
Figure 5—source data 1
AimA reduces neutrophil influx and promotes colonization of Aeromonas.
- https://doi.org/10.7554/eLife.37172.022
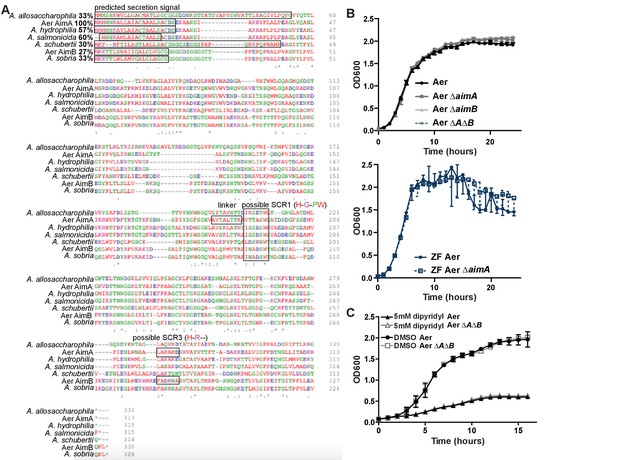
Homologues to AimA are found across the Aeromonas genus.
(A) Clustal Omega alignment of six AimA homologues with AimA. The predicted secretion signal, the linker between the N- and C-term, and two possible lipocalin SCRs are indicated on the alignment. (B) In vitro growth curves of Aer ΔaimA, Aer ΔaimB, and Aer ΔAΔB in A. veronii strain Hm21, and ZF Aer ΔaimA in the zebrafish Aeromonas background do not have growth defects in vitro. (C) In vitro growth curves of wild type Aeromonas (Aer) and Aer ΔAΔB grown in iron limiting conditions (+dipyridyl) or with a vehicle control (DMSO).
-
Figure 5—figure supplement 1—source data 1
Homologues to AimA are found across the Aeromonas genus.
- https://doi.org/10.7554/eLife.37172.023
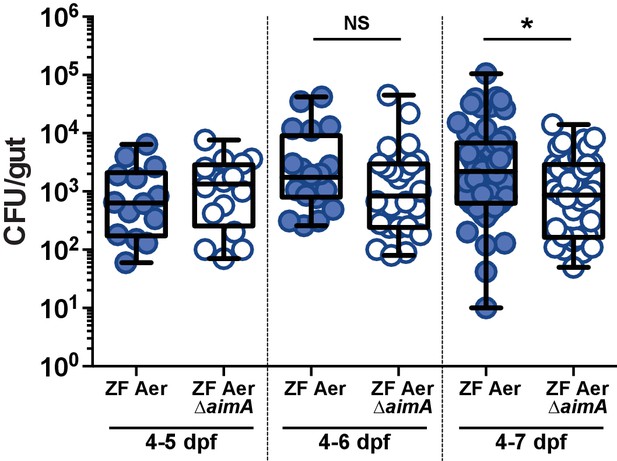
ZF Aer ΔAimA suffers the greatest colonization defect seven dpf.
Comparison of WT ZF Aeromonas (ZOR001) and ZF Aer ΔAimA colonizing the zebrafish intestine for 24 hr (4–5 dpf), 48 hr (4–6 dpf), or 72 hr (4–7 dpf). *p<0.05, Students T test. N > 14, from at least two independent experiments.
-
Figure 5—figure supplement 2—source data 1
ZF Aer DAimA suffers the greatest colonization defect 7 dpf.
- https://doi.org/10.7554/eLife.37172.024
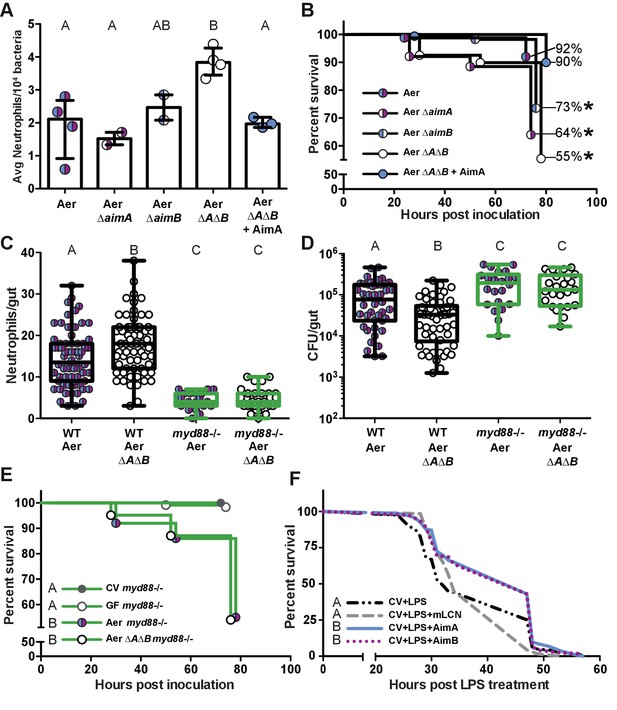
The increased neutrophil response to Aeromonas ΔAΔB causes decreased survival rate.
(A) The per capita effect of wild-type Aeromonas, ΔAimA, ΔAimB, and ΔAΔB. Each dot represents the average neutrophil response from a flask of 15 fish divided by the average colonization level from a flask of 15 fish, normalized to 104. (B) Survival curve of zebrafish mono-associated with wild-type A. veronii (N = 163), ΔAimA (N = 139), ΔAimB (N = 174), ΔAΔB (N = 148), and ΔAΔB + AimA (N = 60). *indicates significant difference from the survival curve with wild-type Aeromonas, Mantel-Cox test. (C) myd88-/- transgenic fish lack a neutrophil response to bacteria. Each dot represents one fish. (D) ΔAΔB colonization is rescued to wild-type colonization levels in myd88-/- transgenic fish. Further, both wild-type Aeromonas and ΔAΔB reach significantly higher colonization levels in the myd88-/- transgenic fish compared to wild-type fish, indicating that the innate immune response limits commensal bacterial growth. (E) Survival curves of myd88-/- zebrafish over a 3 day infection. GF myd88-/- (N = 45), CV myd88-/- (N = 120), Aer myd88-/- (N = 100), ΔAΔB myd88-/- (N = 124). (F) Survival curve of conventionally raised (CV) fish treated with LPS (N = 163), with LPS and AimA (N = 163), with LPS and AimB (N = 121), or with LPS and mLCN (N = 69). Letters next to key indicate significant difference between the survival curves, Mantel-Cox test. For all graphs, letters indicate significance by ANOVA with multiple comparisons.
-
Figure 6—source data 1
The increased neutrophil response to Aeromonas ΔAΔB causes decreased survival rate.
- https://doi.org/10.7554/eLife.37172.026
Tables
Strain table.
https://doi.org/10.7554/eLife.37172.007| Strain | Characteristics | Ref. or source | Manuscript abbreviation |
|---|---|---|---|
| Hm21S | Parent strain, SmR | Graf, 1999 | Aer |
| HE-1095 | Hm21S::interrupted exeM mTn5 KmR SmR | Maltz and Graf, 2011 | Aer ΔT2 |
| HEC-1344 | HE-1095::Tn7 containing TpR exeMN + promoter region | Maltz and Graf, 2011 | Aer ΔT2C |
| ASRC7 | Hm21S aimA::cmR | This study | Aer ΔaimA |
| ASRD5 | Hm21S ΔaimB | This study | Aer ΔaimB |
| ASRD4 | Hm21S aimA::cmR; ΔaimB | This study | Aer ΔAΔB |
| ZOR0001 | Zebrafish Aeromonas isolate | Stephens | ZF Aer |
| ASRC9 | ZOR0001 aimA::cmR | This study | ZF Aer ΔaimA |
Data Collection and Refinement Statistics for Deposited Model.
https://doi.org/10.7554/eLife.37172.008| Data collection | Iodide | Native | ||||
|---|---|---|---|---|---|---|
| space group | P622 | P622 | ||||
| unit cell a, b, c (Å) | 161.4 | 161.4 | 66.6 | 160.5 | 160.5 | 66.2 |
| alpha, beta, gamma (degrees) | 90 | 90 | 120 | 90 | 90 | 120 |
| resolution (Å) | 50.0–2.7 (2.75–2.70) | 41.1–2.30 | ||||
| completeness (%) | 100 (100) | 100 (100) | ||||
| no. unique reflections | 14772 (711) | 22818 (3244) | ||||
| multiplicity | 37.7 (38.1) | 70.2 (71.6) | ||||
| I/sigma> | 44.2 (3.5) | 16.1 (1.4) | ||||
| CC1/2 | 1 | 0.861 | 1.0 (0.9) | |||
| CC1/2 anomolous | 1 | 0.873 | ||||
| R merge (%) | 19.6 (486) | 19.1 (574) | ||||
| Refinement | ||||||
| R work (%) | 17.2 | |||||
| R free (%) | 20.4 | |||||
| no. of molecules in the asymetric unit | 1 | |||||
| no. protein residues | 290 | |||||
| no. of waters | 69 | |||||
| rmsd for lengths (Å) | 0.008 | |||||
| rmsd for angles (deg) | 1.1 | |||||
| Ramachandran plot (%) | ||||||
| preferred | 96.2 | |||||
| allowed | 3.4 | |||||
| outliers | 0.4 | |||||
| Avg. B factor (Å^2) | ||||||
| mainchain* | 81 | |||||
| waters | 75 | |||||
| new PDB entry | 6B7L |
-
* Several loop regions display high mobility but have been modeled due to a visible chain path in the electron density, resulting in an increase in the average observed B-factors of the main chain
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21 DE3 | New England Biolabs | C2527 | |
| Strain, strain background (E. coli) | K12 strain (MG1655) | doi: 10.1128/JB.188.3.928–933.2006 | Dr. Matthew Mulvey, Univ. of Utah | |
| Strain, strain background (Aeromonas veronii) | HM21S; Aer; Aeromonas Hm21; Hm21 | doi:10.1128/AEM.01621–10 | Parent strain, SmR | |
| Strain, strain background (A. veronii) | HE-1095; Aer DT2 | doi:10.1128/AEM.01621–10 | Hm21S::interrupted exeM mTn5 KmR SmR | |
| Strain, strain background (A. veronii) | HEC-1344; Aer DT2C | doi:10.1128/AEM.01621–10 | HE-1095::Tn7 containing TpR exeMN + promoter region | |
| Strain, strain background (A. veronii) | ASRC7; Aer DaimA | this study | Hm21S aimA::cmR | |
| Strain, strain background (A. veronii) | ASRD5; Aer DaimB | this study | Hm21S DaimB | |
| Strain, strain background (A. veronii) | ASRD4; Aer DADB | this study | Hm21S aimA::cmR; DaimB | |
| Strain, strain background (A. veronii) | ZOR0001; ZF Aer | doi:10.1038/ismej.2015.140 | Zebrafish Aeromonas isolate | |
| Strain, strain background (A. veronii) | ZOR0001; ZF Aer DaimA | this study | ZOR0001 aimA::cmR | |
| Genetic reagent (Danio rerio) | AB x Tu strain; wild type zebrafish | UO Zebrafish facility | ||
| Genetic reagent (Danio rerio) | myd88-/- | PMID: 28973938 | ||
| Genetic reagent (Danio rerio) | Tg(BACmpx:GFP) i114; mpx:GFP | doi:10.1182/blood -2006-05-024075 | ||
| Recombinant DNA reagent | pET21B | Genscript | ||
| Chemical compound, drug | LPS, E. Coli O111:B4 | Sigma | L2630 | |
| Peptide, recombinant protein | Lipocalin 2, LCN2, Siderocalin | Biolegend | 588002 | |
| Peptide, recombinant protein | AimA | this study | NCBI hypothetical protein WP_021230730.1 | see Materials and Methods |
| Peptide, recombinant protein | AimB | this study | NCBI hypothetical protein WP_021230165.1 | see Materials and Methods |
| Peptide, recombinant protein | mLCN2 | this study | NCBI gene NM_008491.1 | see Materials and Methods |
Additional files
-
Supplementary file 1
Mass spectrophotometry results comparing cell-free supernatant from the complement of the type II secretion mutant A. veronii strain Hm21 (WT, ∆T2C) compared to the type II secretion mutant (mut, ∆T2).
Mass-spectrophotometry was performed on the CFS from ∆T2C and ∆T2 determined which proteins were enriched in the ΔT2C compared to the ΔT2 strain. This table lists the top 22 proteins that were enriched by greater than 10 counts in the ΔT2C CFS.
- https://doi.org/10.7554/eLife.37172.027
-
Supplemental file 2
Structural homology hits to the N and C terminal domains of AimA.
Structural homology searches performed against the N and C terminal domains of AimA separately revealed that both domains have similarity to proteins in the calycin superfamily, primarily avidins and lipocalins.
- https://doi.org/10.7554/eLife.37172.028
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37172.029





