SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program
Figures
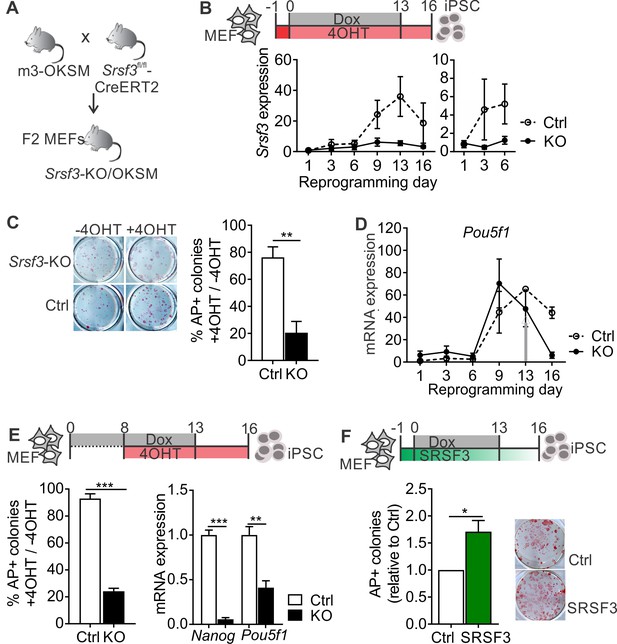
SRSF3 is essential for reprogramming.
(A) The breeding strategy to obtain reprogrammable mice with a conditional Srsf3 knockout allele (Srsf3-KO/OKSM). (B) Experimental outline of the reprogramming experiments (top). Quantification of Srsf3 mRNA levels by RT-qPCR in SRSF3 depleted (KO) and control (Ctrl) cells in the course of reprogramming from day 1 to day 16 (left). Days 1–6 are shown on the right to visualise the expression changes within the first phase of reprogramming. Two-way ANOVA (***p=0.0002 for genotype, ***p=0.0008 for time and *p=0.0154 the interaction, data as mean ± SEM, n = 3–5). (C) Alkaline Phosphatase (AP) labelling of SRSF3 deficient (KO) and control (Ctrl) cells at day 16 of reprogramming (left) and quantification of AP-positive colonies (right). The data is presented as a relative number of colonies between treated (+4 OHT) and untreated (−4OHT) cells reprogrammed in parallel (Unpaired Student’s t-test, two-tailed, **p<0.01, data as mean ± SEM, n = 4–5). (D) Quantification of Pou5f1 mRNA expression by RT-qPCR during reprogramming in SRSF3 depleted (KO) and control (Ctrl) cells. The grey arrow denotes the point of Dox withdrawal and start of endogenous Pou5f1 expression (data as mean ± SEM, n = 2). The data is normalised to Hprt and presented relative to control MEFs. (E) Experimental outline (top) of experiments where MEFs were reprogrammed for 8 days before SRSF3 depletion. Quantification of AP positive colonies (left) and pluripotency marker expression (right) at day 16 of reprogramming as in (C). The AP counts and mRNA expression are presented as above (Unpaired Student’s t-test, two-tailed, **p<0.01, data as mean ± SEM, n = 4). (F) Experimental outline of the SRSF3 overexpression experiments (top). AP labelling of colonies at day 16 of reprogramming and quantification of AP-positive colonies. The data is presented relative to GFP-only (Ctrl) transfected cells (Unpaired Student’s t-test, two-tailed, *p<0.05, data as mean ± SEM, n = 3). See also Figure 1—figure supplement 1.
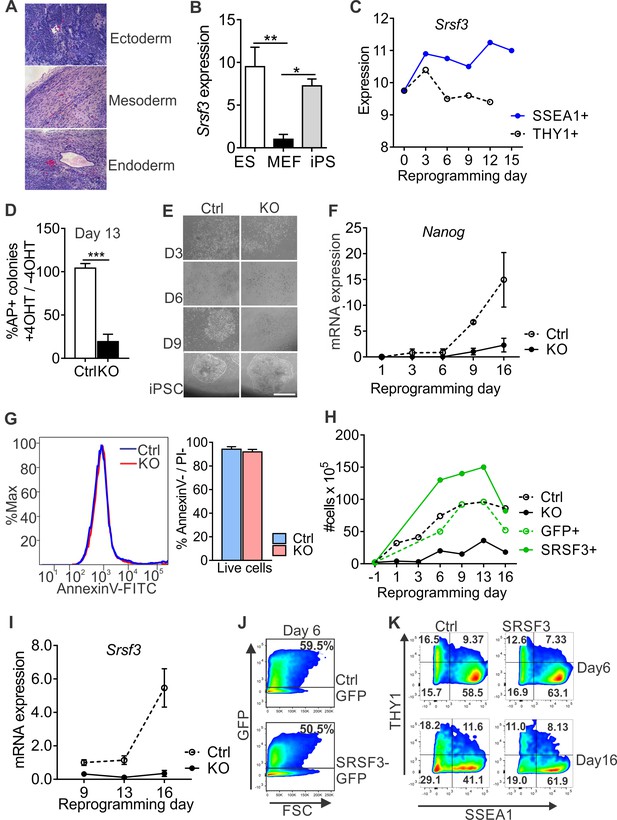
SRSF3 depletion and overexpression have the opposite effects on reprogramming outcome.
(A) iPSCs cells generated from Cre-negative m3-OKSM MEFs form teratomas when injected into immunocompromised mice. (B) Quantification of Srsf3 mRNA levels by RT-qPCR in ESCs, MEFs and iPSCs. One-way ANOVA, Tukey’s multiple comparison test (*p<0.05; **p<0.01, data as mean ± SEM, n = 3). (C) Srsf3 mRNA expression increases during reprogramming in the SSEA1 +population. The graph is based on data from Polo et al. (2012). (D) Quantification of AP-positive colonies in Srsf3-KO/OKSM and control cultures at day 13 of reprogramming. The data is presented as relative number of colonies between treated (+4 OHT) and untreated (−4OHT) samples that were reprogrammed in parallel (Unpaired Student’s t-test, two-tailed, ***p<0.001, data as mean ± SEM, n = 4–5). (E) Phase contrast images of reprogramming Srsf3-KO/OKSM and control cultures at different time points. Scale bar 50 μm. (F) Quantification of Nanog mRNA expression as in Figure 1D. (G) Flow cytometric quantification of apoptotic and dead cells by AnnexinV/PI labelling 48 hr after 4OHT induction in reprogramming Srsf3-KO/OKSM and control MEFs. Histogram of a representative experiment (left) and quantification of live cells (right) are presented (Unpaired Student’s t-test, two-tailed, *p>0.05, data as mean ± SEM, n = 3). (H) Quantification of cell numbers during reprogramming in control (Ctrl), Srsf3-KO/OKSM (KO), GFP-only control (GFP+) and SRSF3-T2A-GFP overexpressing (SRSF3+) cells (data as mean). (I) Quantification of Srsf3 mRNA expression after 4OHT induction at day 8 as in (F). Data is presented relative to day 9. (J) Flow cytometric analysis of GFP and SRSF3-T2A-GFP expression on day 6 of reprogramming. (K) Analysis of SSEA1 and THY1 cell surface marker expression at days 6 and 16 of reprogramming in GFP-only (Ctrl) and SRSF3-T2A-GFP (SRSF3) overexpressing cells. Live GFP+ cells were gated and cell surface markers assessed in the transduced cell population.
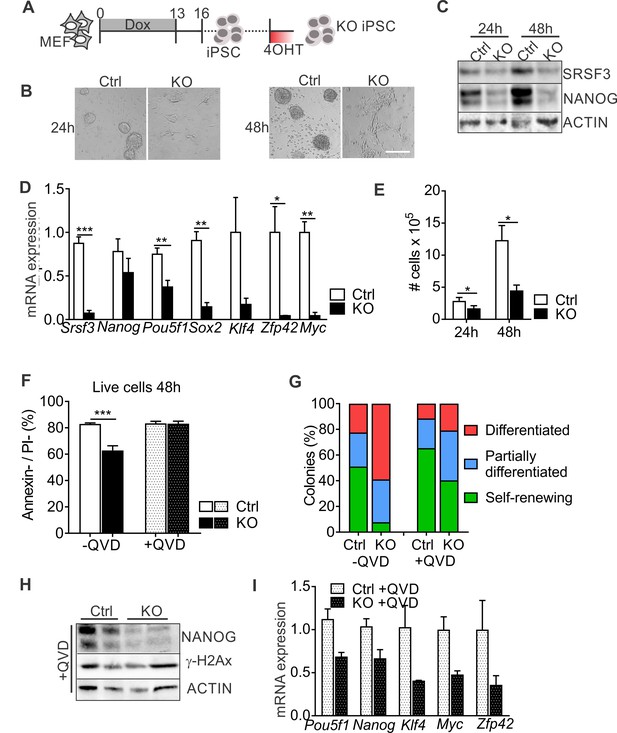
SRSF3 depletion leads to loss of pluripotency independent of cell death.
(A) Experimental outline of the Srsf3-KO iPSC generation. (B) Phase contrast images of SRSF3 depleted (KO) and control (Ctrl) iPSCs 24 and 48 hr after 4OHT induction. Scale bar: 50 μm. (C) Western blot analysis of SRSF3 and NANOG expression in SRSF3 deficient (KO) and control (Ctrl) iPSCs 24 and 48 hr after 4OHT induction. ACTIN served as a loading control. (D) Quantification of Srsf3 and pluripotency marker mRNA expression in Srsf3-KO (KO) and control (Ctrl) iPSCs 24 hr after 4OHT induction. The data is normalised to Hprt and presented relative to control iPSCs (Unpaired Student’s t-test, two-tailed, *p<0.05, **p<0.01, ***p<0.001, data as mean ± SEM, n = 3). (E) Quantification of cell numbers 24 and 48 hr after 4OHT induction in Srsf3-KO (KO) and control (Ctrl) iPSCs (Unpaired Student’s t-test, two-tailed, *p<0.05, data as mean ± SEM, n = 4). (F) Quantification of live cells in untreated and QVD treated Srsf3-KO (KO) and control (Ctrl) iPSCs by AnnexinV/PI labelling and flow cytometry 48 hr after 4OHT induction (Unpaired Student’s t-test, two-tailed, *p<0.05, data as mean ± SEM, n = 3). (G) Quantification of pluripotent and differentiated iPSC colonies in untreated and QVD treated Srsf3-KO (KO) and control (Ctrl) iPSCs. Colonies were scored from three independent experiments. (H) Western blot analysis of γH2Ax and NANOG expression in QVD treated Srsf3-KO (KO) and control (Ctrl) iPSCs 48 hr after 4OHT induction. ACTIN served as a loading control. (I) Quantification of Srsf3 and pluripotency marker mRNA expression in QVD treated Srsf3-KO (KO) and control (Ctrl) iPSCs 24 hr after 4OHT induction. The data is normalised to Hprt and presented relative to control iPSCs (data as mean ± SEM, n = 2). See also Figure 2—figure supplement 1.
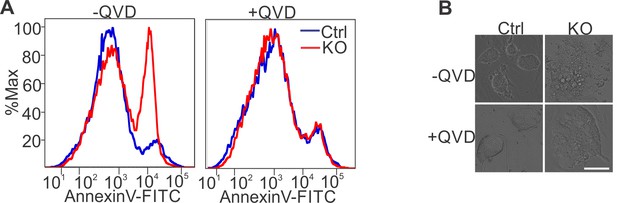
Apoptosis inhibition in SRSF3 depleted iPSCs.
(A) Histogram of a representative experiments of data presented in Figure 2F. (B) Phase contrast images of untreated and QVD treated Srsf3-KO and control iPSCs 48 hr after 4OHT induction. Scale bar: 50 μm.
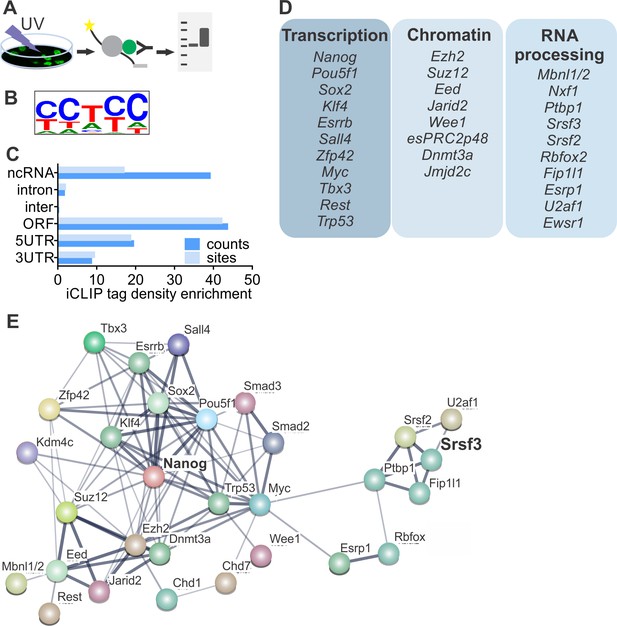
SRSF3 binds to RNAs encoding key pluripotency factors.
(A) Schematic of iCLIP to determine SRSF3 binding sites in pluripotent cells. (B) SRSF3 consensus binding motif in pluripotent cells. (C) Distribution of significant SRSF3 crosslink sites (FDR < 0.05) over different transcript regions normalised to feature length. (D) SRSF3 binding sites detected in RNAs encoding key pluripotency regulators of multiple functional categories. (E) Functional association network of SRSF3 and its RNA targets in pluripotent cells. No connection between SRSF3 and the NANOG centred pluripotency network has been previously assigned. Pluripotency regulators that were directly bound by SRSF3 were used as input for STRING (Szklarczyk et al., 2015). The pluripotency associated transcription factors and chromatin modifiers were based on Young (2011) and mRNA processing factors based on Chen and Hu (2017). The thickness of the line reflects the confidence level for the interaction (highest 0.9 – high 0.7 – medium 0.4 – low 0.15), large nodes represent proteins with some structural information. See also Figure 3—figure supplement 1 and Figure 3—source data 1–4.
-
Figure 3—source data 1
iCLIP peaks for SRSF3-EGFP.
- https://doi.org/10.7554/eLife.37419.008
-
Figure 3—source data 2
iCLIP peaks for EGFP-NLS.
- https://doi.org/10.7554/eLife.37419.009
-
Figure 3—source data 3
iCLIP peaks for anti-SRSF3.
- https://doi.org/10.7554/eLife.37419.010
-
Figure 3—source data 4
SRSF3 iCLIP mapping statistics.
- https://doi.org/10.7554/eLife.37419.011
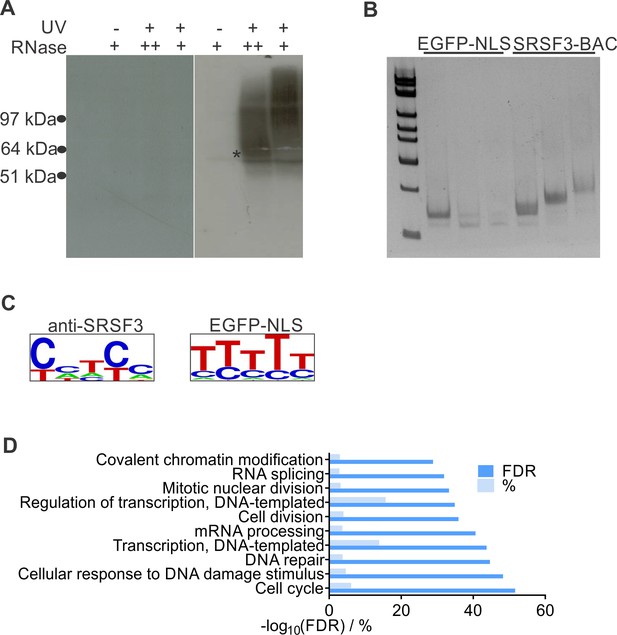
SRSF3 iCLIP in pluripotent cells.
(A) Protein-RNA complexes isolated for SRSF3-GFP and control iCLIP library preparation. The star marks the position of SRSF3-GFP. The iCLIP library was isolated from a membrane piece above the star for both SRSF3-GFP and GFP-NLS. (B) Amplified iCLIP libraries after size selection. The two largest size fractions were pooled and sequenced, the smallest fraction omitted. (C) SRSF3 consensus binding motif determined from iCLIP conduction using anti-SRSF3 antibody in wild type (left) and GFP-antibody in GFP-only expressing (right) pluripotent cells. (D) Analysis of enriched GO terms among RNAs with significant SRSF3 crosslink sites.
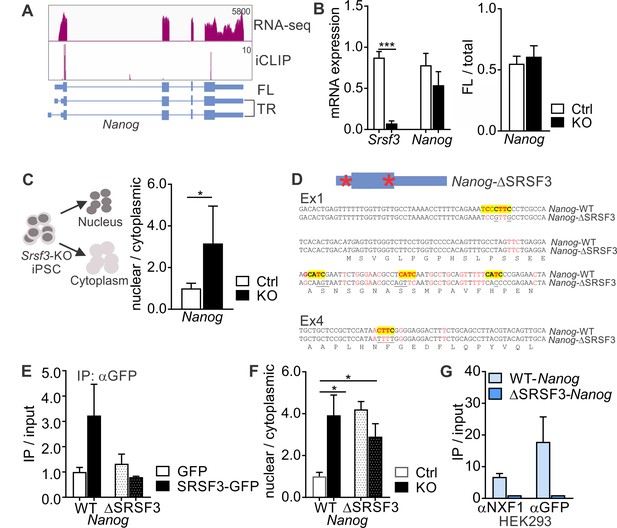
SRSF3 binding and mRNA export adaptor activity in independent of splicing and essential for Nanog mRNA export.
(A) Nanog expression and SRSF3 iCLIP binding profile within Nanog transcript region. Gene diagram depicting Nanog variants is presented below the graphs (FL = full length, TR = truncated). (B) Left: RT-qPCR quantification of Srsf3 and Nanog mRNA expression in Srsf3-KO (KO) and control (Ctrl) iPSCs 24 hr after 4OHT induction. The data is normalised to Hprt and presented relative to control iPSCs (***p<0.001, Unpaired Student’s t-test, two-tailed, data as mean ± SEM, n = 3). Right: Quantification of FL Nanog transcript levels relative to total Nanog mRNA levels (p>0.05, Unpaired Student’s t-test, two-tailed, data as mean ± SEM, n = 3). (C) Srsf3-KO (KO) and control (Ctrl) iPSCs were fractionated into nuclear, cytoplasmic and total fractions 24 hr after 4OHT induction (left). Quantification of Nanog mRNA levels in the nuclear and cytoplasmic fractions (right). Data is presented as nuclear to cytoplasmic ratio (*p<0.05, Unpaired Student’s t-test, two-tailed, data as mean ± SEM, n = 5) and normalised to control. #error bar smaller than the border line. (D) Generation of WT-Nanog and ΔSRSF3-Nanog expression constructs. Both constructs contain the complete Nanog cDNA, including 5’ and 3’ UTRs. In ΔSRSF3-Nanog, synonymous mutations have been introduced to remove SRSF3 binding sites (positions within the transcript denoted by red asterisks in the gene diagram). SRSF3 crosslink nucleotides are shown in red, SRSF3 consensus sequences are highlighted in yellow, and the underlined nucleotides were mutated to abolish SRSF3 consensus binding sites without affecting the coding potential. (E) RNA immunoprecipitation (IP) of WT- and ΔSRSF3-Nanog mRNA in SRSF3-GFP or GFP-only expressing pluripotent cells. The Nanog mRNA enrichment is presented as IP/input and normalised to GFP-only control cells expressing WT-Nanog (mean ± SEM, n = 2). (F) Srsf3-KO (KO) and control (Ctrl) iPSCs expressing WT-Nanog or ΔSRSF3-Nanog were fractionated into nuclear, cytoplasmic and total fractions 24 hr after 4OHT. Quantification of WT-Nanog and ΔSRSF3-Nanog mRNA levels in the nuclear and cytoplasmic fractions is presented as nuclear to cytoplasmic ratio and normalised to the control cells expressing WT-Nanog (*p<0.05, One-way ANOVA, data as mean ± SEM, n = 3). (G) RNA immunoprecipitation of WT- and ΔSRSF3-Nanog mRNA in SRSF3-GFP or GFP-only expressing HEK293 cells. Anti-NXF1 and anti-GFP antibodies were used to enrich mRNAs bound by NXF1 and SRSF3-GFP, respectively. The Nanog mRNA enrichment is presented as IP/input and normalised to GFP-only cells expressing WT-Nanog (mean ± SEM, n = 2). See also Figure 4—figure supplement 1.
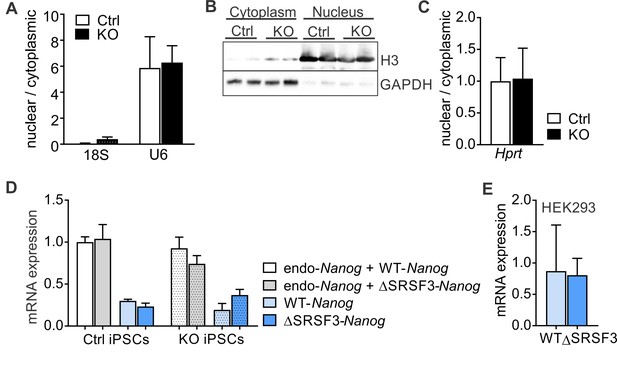
SRSF3 binding is essential for Nanog mRNA export.
(A) Srsf3-KO and control iPSCs were fractionated into nuclear and cytoplasmic fractions 24 hr after 4OHT induction. RT-qPCR quantification of cytoplasmic 18S ribosomal RNA and nuclear U6 snRNA levels in the nuclear and cytoplasmic fractions. Data is presented as nuclear to cytoplasmic ratio (data as mean ± SEM, n = 2). (B) Western blot analysis of the nuclear histone H3 and cytoplasmic GAPDH in the subcellular fractions of control and Srsf3-KO iPSCs. (C) RT-qPCR quantification of Hprt mRNA in the nuclear and cytoplasmic fractions Srsf3-KO and control iPSCs, showing no change in Hprt distribution following SRSF3 depletion. Data is presented as nuclear to cytoplasmic ratio (data as mean ± SEM, n = 5). (D) RT-qPCR quantification ofa endogenous Nanog mRNA expression and the ectopically expressed WT-Nanog and ΔSRSF3-Nanog mRNAs in control and SRSF3 depleted pluripotent cells (data as mean ± SEM, n = 3, ns). (E) RT-qPCR quantification of ectopically expressed WT-Nanog and ΔSRSF3-Nanog mRNAs in HEK293 cells (data as mean ± SEM, n = 2).
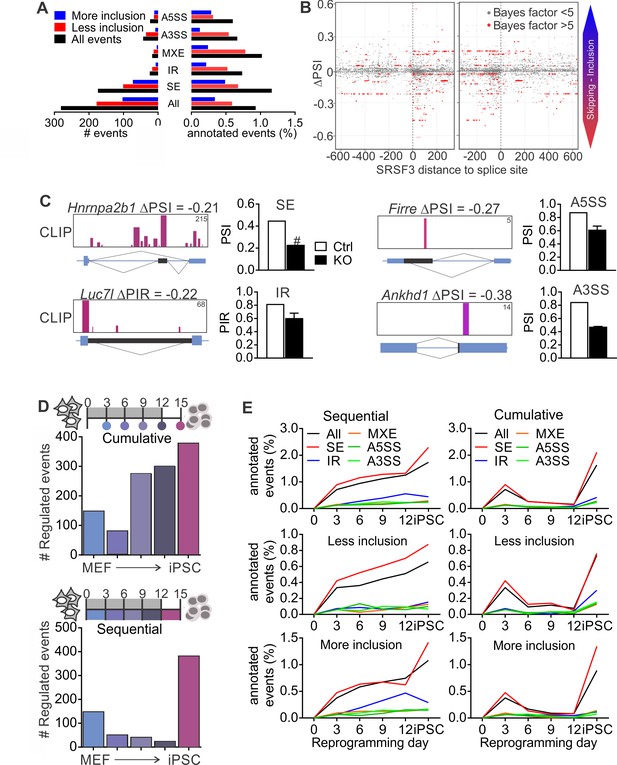
SRSF3 regulated alternative splicing in pluripotent cells and the dynamics of AS during reprogramming.
(A) Different alternative splicing (AS) types affected by SRSF3 depletion in pluripotent cells. Pluripotent cells were subjected to RNA sequencing 24 hr following SRSF3 depletion and AS changes were quantified by MISO (ΔPSI ≥ |0.2| and Bayes factor ≥5). SE, skipped exon; IR, intron retention; MXE, mutually exclusive exon; A3SS, alternative 3’ slice site; A5SS, alternative 5’ splice site. (B) SRSF3 binding sites (iCLIP) at and around SE events that were annotated by MISO. The red dots represent AS events that were considered significantly changed following SRSF3 depletion (Bayes factor >5) and grey dots events that did not change (Bayes factor <5). The opacity of the dots corresponds to the log2(iCLIP count) in the range 1 to 5 before log transformation. Significant SRSF3-binding sites with iCLIP count ≥2 that were not detected in the control iCLIP are shown. The dotted lines at 0 denote the 3’ and 5’ splice sites. (C) Representative examples of different AS types affected by SRSF3 depletion. SRSF3-binding sites mapping at and around the affected event are shown over the gene diagram (PSI values based on MISO, data as mean ± SEM, n = 2). PSI, percent spliced in; PIR, percent intron retained. Ctrl, control cells; KO, SRSF3-depleted cells. (D) Dynamic AS changes take place during reprogramming (MISO, ΔPSI ≥ |0.2| and Bayes factor ≥5). Comparison of each time point to MEFs showing a cumulative increase in AS from MEFs to iPSC (top) and assessment of consecutive time points demonstrating that the majority of AS changes take place in only two time points (below). (E) Different AS types display distinct dynamics during reprogramming. Cumulative and sequential change as in D, event types as in A. See also Figure 5—figure supplement 1 and Figure 5—source data 1–3.
-
Figure 5—source data 1
Alternative splicing events following SRSF3 depletion.
- https://doi.org/10.7554/eLife.37419.016
-
Figure 5—source data 2
Alternative splicing events during reprogramming relative to MEFs.
- https://doi.org/10.7554/eLife.37419.017
-
Figure 5—source data 3
Alternative splicing events between consecutive time points during reprogramming.
- https://doi.org/10.7554/eLife.37419.018
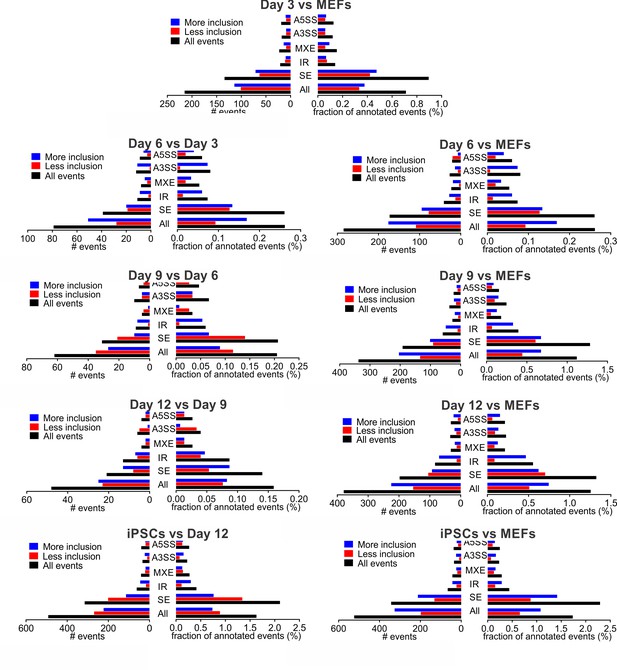
Alternative splicing during reprogramming.
Changes in different alternative splicing (AS) types during reprogramming. AS changes in each of the reprogramming intermediates were quantified by MISO (ΔPSI ≥ |0.2| and Bayes factor ≥5) either by comparing to the previous time point (left, sequential) or MEFs (right, cumulative). SE, skipped exon; IR, intron retention; MXE, mutually exclusive exon; A3SS, alternative 3’ slice site; A5SS, alternative 5’ splice site.
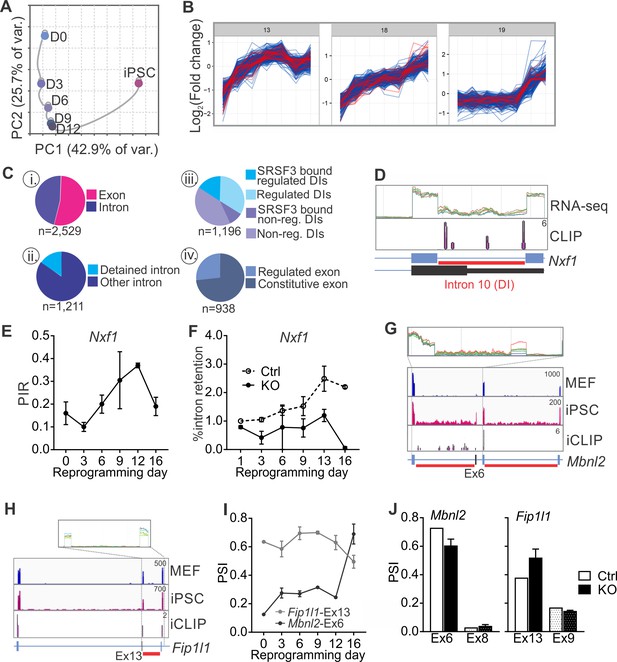
SRSF3 regulates the alternative splicing of Nxf1 and other RNA regulators during reprogramming.
(A) Principal component analysis of regulated RNA features based on JunctionSeq during reprogramming, depicting similarity of exon/junction usage between reprogramming intermediates. (B) Unsupervised k-means clustering of regulated RNA features during the reprogramming time course. SRSF3 associated regulated RNA features, SARFs, are highlighted in red. SRSF3 binding sites within the feature or [−200, +200 nt] window up- or down-stream of the junction were included in the analysis. Top three clusters with most SRSF3-binding sites are shown. (C) (i) Distribution of SRSF3-binding sites within regulated exons and introns. Regulated RNA features were classified as exons and introns using RefSeq annotation. (ii) Proportion of intronic SARFs that were classified as detained introns (Boutz et al., 2015). (iii) Distribution of SRSF3 binding sites within regulated and non-regulated (non-reg.) DIs. (iv) Proportion of SARFs that are DIs and flank an exon regulated during reprogramming. (D) Expression of Nxf1 Ex10-Int10-Ex11 during reprogramming and SRSF3 iCLIP binding profile within the transcript region. The red bars below the gene diagram denote DIs based on (Boutz et al., 2015). Colour coding from blue (MEF) to red (iPSC). The read counts were normalised within the genes. (E) Dynamic changes in Nxf1-Int10 retention during reprogramming as measured by RNA-seq (data as mean ± SEM, n = 2). PIR, percent intron retained. (F) RT-qPCR quantification of Nxf1-Int10 retention during reprogramming in control (Ctrl) and SRSF3-depleted (KO) cells. The data is represented as percent of transcripts with the retained intron (data as mean ± SEM, n = 2). (G) Expression of Mbnl2 exons 5–8 and SRSF3 iCLIP binding profile within the transcript region. The red bars below the gene diagram denote DIs based on (Boutz et al., 2015). The alternative exon six is highlighted in black in the gene diagram. Colour coding from blue (MEF) to red (iPSC). The read counts were normalised within the genes. (H) Expression of Fip1l1 exons 12–14 and SRSF3 iCLIP binding profile within the transcript region. The red bars below the gene diagram denote DIs based on (Boutz et al., 2015). The alternative exon 13 is highlighted in black in the gene diagram. (I) Dynamic changes in Mbnl2-Ex6 and Fip1l-Ex13 inclusion during reprogramming as measured by RNA-seq (data as ± SEM, n = 2). PSI, precent spliced in. (J) Quantification of Mbnl2-Ex6 and Fip1l-Ex13 inclusion in control (Ctrl) and Srsf3-KO (KO) iPSCs by RNA-seq (data as mean ± SEM, n = 2). Unaffected alternative exons that were not associated with SRSF3 binding sites (Mbnl2- Ex8 and Fip1l1-Ex9) are shown for comparison. PSI, percent spliced in. See also Figure 6—figure supplement 1, Figure 6—source data 1 and 2.
-
Figure 6—source data 1
Read-wide mapping statistics.
- https://doi.org/10.7554/eLife.37419.021
-
Figure 6—source data 2
Data file containing JunctionSeq output.
- https://doi.org/10.7554/eLife.37419.022
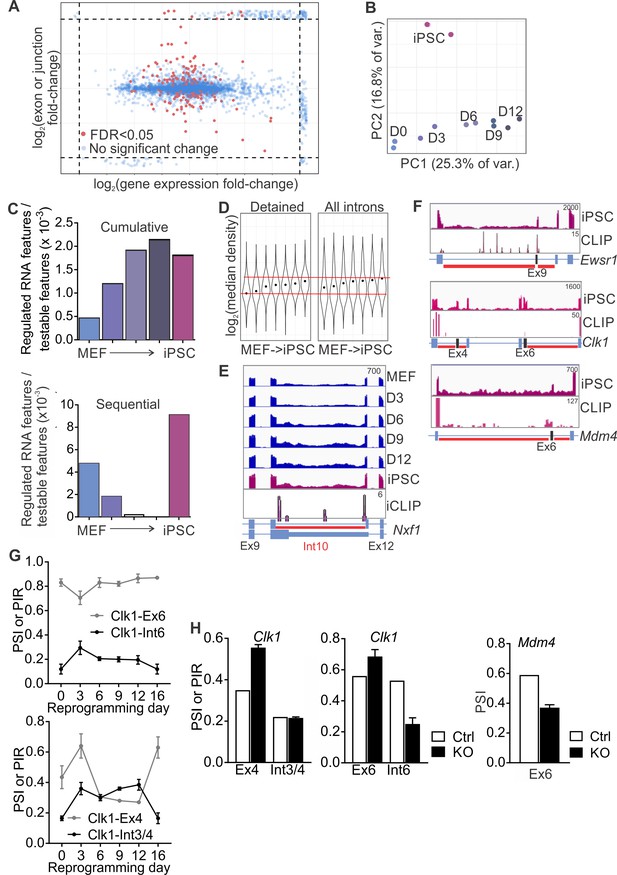
SRSF3 controls the alternative splicing of RNA regulators.
(A) Alternative splicing changes during reprogramming as determined by JunctionSeq are independent of mRNA expression levels. Five-thousand features were randomly chosen from the JunctionSeq output (Figure 6—source data 2); where a fold-change fell outside the plots range or was infinite, for instance where the denominator read-count was zero, it was plotted in the margins (dashed lines). (B) Principal component analysis of regulated RNA features during reprogramming, with individual replicates shown. (C) Frequency of regulated RNA features during the reprogramming time course when each time point was compared to MEFs (cumulative) or to the previous time point (sequential). The data is presented as the number of regulated RNA features relative to all testable RNA features within each comparison. (D) Violin plots depicting distribution of expression density (JunctionSeq expression estimates divided by intron length) for introns > 100 nt, in genes containing detained introns. Gene-wise medians for detained introns or other introns are shown. Dots indicate geometric mean of each time-point. (E) RNA-seq read profiles of Nxf1 gene regions around a detained intron, depicting the dynamic regulation of the intron inclusion during reprogramming. (F) Expression of Ewsr1, Clk1 and Mdm4 at and around DI regions in iPSCs and SRSF3 iCLIP binding profiles within the transcript regions. The red bars below gene diagram denote detained introns based on a previous study (Boutz et al., 2015). The alternative exon is marked in black in the gene diagram. (G) Dynamic regulation of Clk1 alternative exons 4 and 6 and detained introns 3/4 and 6 during reprogramming. PSI, percent spliced in and PIR, percent intron retained (RNA-seq data as mean ± SEM, n = 2). (H) RNA-seq quantification of Clk1 and Mdm4 exon inclusion and intron retention following SRSF3 depletion iPSCs (data as mean ± SEM, n = 2). PSI, percent spliced in and PIR, percent intron retained.
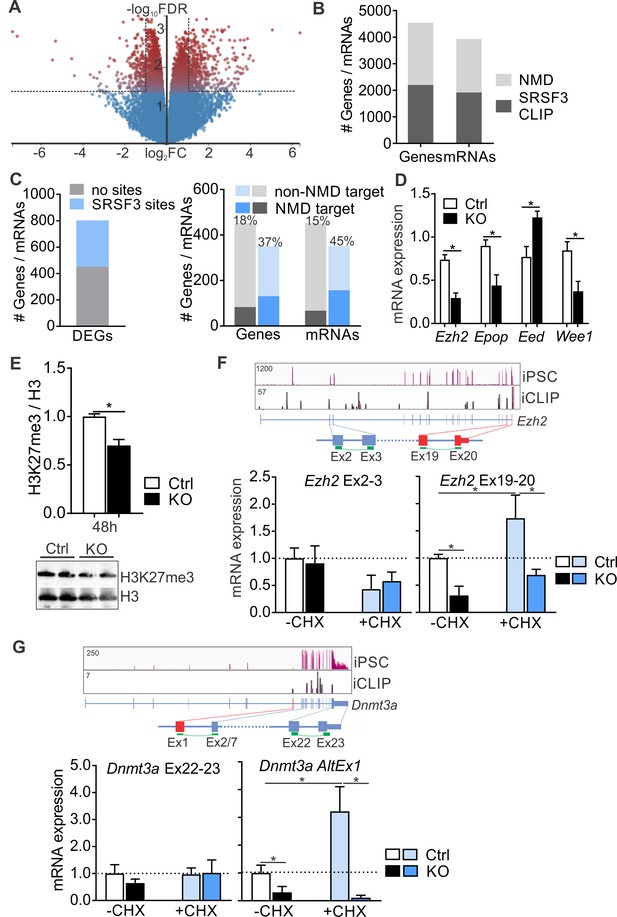
SRSF3 mediates the abundance of distinct mRNAs encoding chromatin modifiers to facilitate pluripotency.
(A) Volcano plot depicting differentially expressed genes following SRSF3 depletion in pluripotent cells. The dotted lines illustrate fold change (FC) >2 and FDR < 0.05. (B) Number of genes and mRNAs regulated at the level of NMD based on Hurt et al. (2013) and the fraction of them with significant SRSF3 crosslink sites. (C) Left: Differentially expressed genes (DEGs) following SRSF3 depletion with and without SRSF3-binding sites. Right: The fraction of differentially expressed genes/mRNAs with (blue bars) and without (grey bars) SRSF3-binding sites that were regulated at the level of NMD. The percent value above the bars denotes the fraction of NMD regulated genes or mRNAs (dark blue or grey) in each category. (D) RT-qPCR quantification of PRC2 components in Srsf3-KO (KO) and control (Ctrl) iPSCs 24 hr after 4OHT induction. The data is normalised to Hprt and presented relative to control iPSCs (*p<0.05, Unpaired Student’s t-test, two-tailed, data as mean ± SEM, n = 3–9). (E) Western blot analysis of H3K27me3 and total H3 levels 48 hr after 4OHT induction in Srsf3-KO (KO) and control (Ctrl) iPSCs. Quantification presented as H3K27me3 relative to total H3 (*p<0.05, Unpaired Student’s t-test, two-tailed, data as mean ± SEM, n = 2). (F) Top: Ezh2 expression in iPSCs and SRSF3 iCLIP binding profiles within the transcript region. Primers binding to the exons in red were used to measure the NMD-regulated transcript variant. Below: RT-qPCR quantification of total Ezh2 (Ex2-3) and full-length transcript variant (Ex19-20) in control (Ctrl) and Srsf3-KO (KO) iPSCs before (-CHX) and after (+CHX) NMD inhibition. The data is normalised to Hprt and presented relative to control iPSCs (*p<0.05, One-way ANOVA, data as mean ± SEM, n = 3). (G) Top: Dnmt3a expression in iPSCs and SRSF3 iCLIP binding profiles within the transcript region. Primers detecting the alternative first exon (red) were used to measure the NMD-regulated transcript variant. Below: RT-qPCR quantification of total Dnmt3a (Ex22-23) and Dnmt3a alternative exon 1 (AltEx1) containing transcripts as in (F). See also Figure 7—source data 1.
-
Figure 7—source data 1
Differentially expressed genes following SRSF3 depletion.
- https://doi.org/10.7554/eLife.37419.025
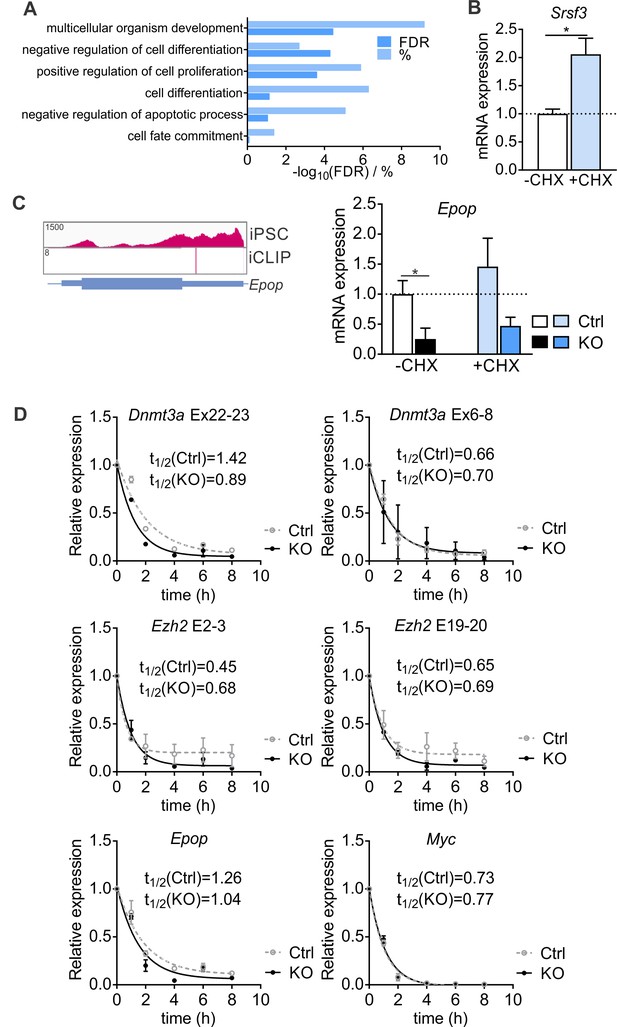
SRSF3 controls the abundance of chromatin modifiers.
(A) Enriched GO terms of differentially expressed genes following SRSF3 depletion. (B) Srsf3 mRNA levels before (-CHX) and after (+CHX) NMD inhibition (Unpaired Student’s t-test, two-tailed, *p>0.05, data as mean ± SEM, n = 3). (C) Expression of Epop in iPSCs and SRSF3 iCLIP binding profile within the transcript region. RT-qPCR quantification of Epop mRNA in control and Srsf3-KO iPSCs before (-CHX) and after (+CHX) NMD inhibition. The data is normalised to Hprt and presented relative to control iPSCs (*p<0.05, One-way ANOVA, data as mean ± SEM, n = 3). (D) Determination of mRNA half-lives in cells treated with Actinomycin D. SRSF3 depletion was induced with 4OHT and 24 hr later Actinimycin was added (t = 0). mRNA abundance was measured by RT-qPCR at the indicated time points and mRNA half-lives determined by fitting the data to nonlinear one phase decay (data as mean ± SEM, n = 2).
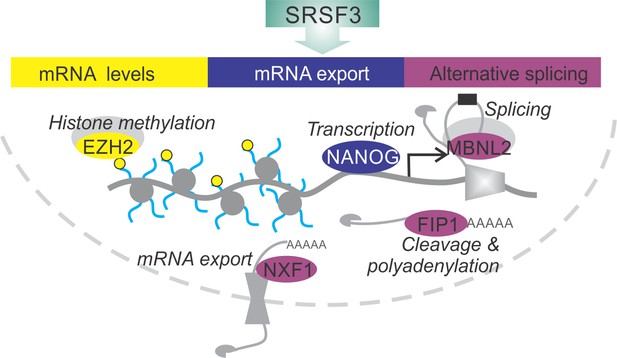
SRSF3 drives self-renewal by regulating RNAs encoding key pluripotency factors.
SRSF3 directly regulates the core pluripotency network through its activities in mRNA export, alternative splicing and mRNA turnover. Through the control of RNAs encoding chromatin modifiers, transcription and RNA processing factors that are involved in the establishment and maintenance of pluripotency SRSF3 has a broad impact on the pluripotency gene expression program.
Tables
Primers used in this study.
F = forward primer, R = reverse primer.
| Gene symbol | Sequence | |
|---|---|---|
| Srsf3 | F | TGAATTAGAACGGGCTTTTGG |
| R | TTCACCATTCGACAGTTCCAC | |
| Hprt | F | TGTTGTTGGATATGC |
| R | TGCGCTCATCTTAGG | |
| Nanog | F | ACCAAAGGATGAAGTGCAAGC |
| R | TGGATGCTGGGATACTCCACT | |
| Nanog TR | F | GCAGTTTTTCATCCCGAGAAC |
| R | GAAGAGGCAGGTCTTCAGAGG | |
| Nanog FL | F | TGACATGAGTGTGGGTCTTCC |
| R | GAAGAGGCAGGTCTTCAGAGG | |
| Pou5f1 | F | GAGGAAGCCGACAACAATGAG |
| R | 5'ATCTGCTGTAGGGAGGGCTTC | |
| Sox2 | F | GTAAGATGGCCCAGGAGAACC |
| R | ATAATCCGGGTGCTCCTTCAT | |
| Klf4 | F | GAAAAGAACAGCCACCCACAC |
| R | CCTGTCACACTTCTGGCACTG | |
| Zfp42 | F | AGATTAGCCCCGAGACTGAGG |
| R | AAGGGAACTCGCTTCCAGAAC | |
| Myc | F | CTGTACCTCGTCCGATTCCAC |
| R | GGTTTGCCTCTTCTCCACAGA | |
| Nxf1 | F | TTCTGCCTGTCTGTTGTCTCC |
| R | CAGAACAGAAAAGGGGAGGTG | |
| Mbnl2 | F | AAAGCACTGAAGCGACCTCTC |
| R | AGAGCCTGCTGGTAGTGCAAG | |
| Ewsr1 | F | GCTTCAATAAGCCTGGTGGAC |
| R | TGCCAGATCATCCAGAGTCAC | |
| Fil1l1 | F | TCCAATAACTGTACCACCTCCA |
| R | CCATAGGGAACGCTCGTG | |
| Ezh2 Ex2-3 | F | AATCTGAGAAGGGACCGGTTT |
| R | ATGTGCACAGGCTGTATCCTC | |
| Ezh2 Ex19-20 | F | GGGCTATCCAGACTGGTGAAG |
| R | CCTGAAGCTAAGGCAGCTGTT | |
| Epop | F | CCGGCTGATGCTCTTTCTACT |
| R | CCGCTAAACTGACCCTCATTC | |
| Eed | F | GGCAAACTGTATGTTTGGGATT |
| R | TCGCAGACAGCTATGAGGATG | |
| Wee1 | F | GAGCTGGTGAAGCATTCAGTG |
| R | CATCCGATCTGTGAAGAGTGC | |
| Dnmt3a Ex22-23 | F | GGGGACCCCTACTACATCAGC |
| R | AGAGGCCTGGTTCTCTTCCAC | |
| Dnmt3a AltEx1 | F | CCAGACGGGCAGCTATTTACA |
| R | AGAGGCCTGGTTCTCTTCCAC | |
| Nanog-WT and Nanog-ΔSRSF3 | F | CAAGCCTCAGACAGTGGTTCA |
| R | ATGTCAGTGTGATGGCGAGG | |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.37419.028





