Optogenetic dissection of mitotic spindle positioning in vivo
Figures
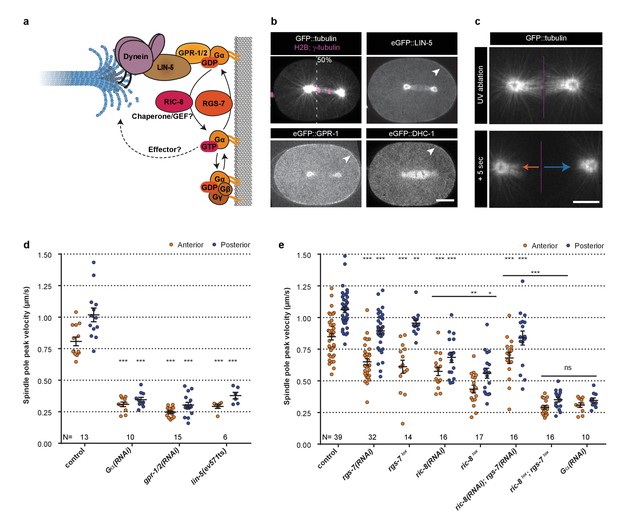
Gα regulation by RIC-8 and RGS-7 is essential for cortical pulling force generation.
(a) Cartoon model representing mechanisms and functions discussed in the text. Gα∙GDP–GPR-1/2–LIN-5–dynein anchors dynamic microtubule plus-ends and generates cortical pulling forces on the mitotic spindle. Gα∙GDP can assemble a Gαβγ or Gα–GPR-1/2–LIN-5 trimer. The Gα∙GDP/GTP nucleotide state is regulated by the GAP RGS-7. For RIC-8, functions as Gα GEF and chaperone are reported. Gα∙GTP could promote spindle positioning through unknown downstream effectors. (b) Spinning disk confocal images of anaphase spindle positioning away from the cell center (dashed line) in the C. elegans zygote. The upper left panel shows the spindle with labeled tubulin and DNA. Other panels: endogenous GPR-1, LIN-5, and dynein (DHC-1) fused to eGFP are present in the cytoplasm, at the cell cortex (arrowheads), and spindle structures. Scale bar: 10 µm. (c) Spinning disk confocal images of the mitotic spindle (marked by GFP::tubulin). Upon UV-laser ablation of the spindle midzone (violet line), spindle poles separate with velocities that represent the respective net force acting on each pole (arrows). Scale bar: 5 µm. (d) Spindle pole peak velocities after midzone ablation. Control is the gfp::tubulin strain. Other conditions: inactivation of Gα, GPR-1/2, and LIN-5. Error bars: s.e.m. Welch’s Student’s t-test; ***p<0.001. (e) Spindle severing experiments in embryos where RIC-8 and RGS-7 were depleted by RNAi or induced tissue specific CRE-lox-mediated knockout of the endogenous gene (lox). Control is the gfp::tubulin strain, see Figure 1—figure supplement 1 for knockout method and additional controls. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; *p<0.05, **p<0.01, ***p<0.001. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
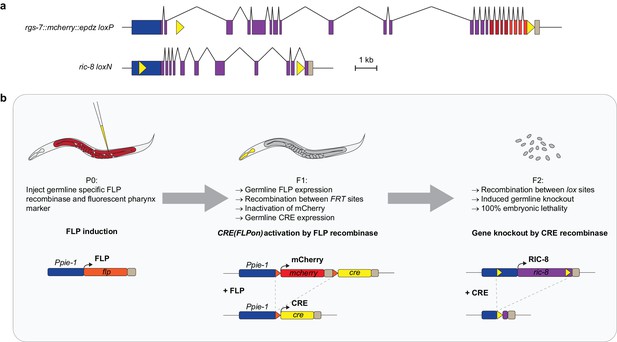
Method of inducible knockout of essential genes in the C. elegans germline.
(a) Position of lox sites (yellow triangles) in rgs-7 and ric-8 loci with promoter (blue), coding sequence (purple), 3’ UTR (khaki), mcherry (red), and epdz (orange). We paid attention not to disturb annotated genetic elements or to create splice donor/acceptor consensus sites. Scale bar: 1 kb. (b) Flow chart of the inducible tissue-specific knock-out procedure. The system depends on successive expression of two recombinases: FLP and CRE. This bipartite nature allows for both spatial and temporal control. For spatial control, we chose the long Ppie-1 germline-specific promoter including enhancer, which is active in every region of the germline during all developmental stages (Merritt et al., 2008). Temporal control depends on injection of the Ppie-1::flp construct (including a fluorescent co-injection marker) in P0 germlines. Transgenic F1 express germline FLP, which recombines between two FRT sites (orange triangles). As a result, the integrated tissue-specific CRE(FLPon) construct switches expression from mcherry to cre. mcherry and cre sequences were germline-optimized for reliable germline expression (see Figure 2—figure supplement 1,2). For the CRE(FLPon) construct, we chose the cep-1 3' UTR because it contains a strong poly-adenylation signal consensus (aataaa) and permits expression in every region and developmental stage of the germline (Merritt et al., 2008). The induced, tissue-specific CRE recombines between lox sites (yellow triangles), generating a knockout and 100% embryonic lethal F2 generation. The two-step knockout creates a delay that results in germline CRE activity only in the F1 germline. P0 germline CRE activity generates knockout F1 animals but does not deplete maternal protein product. See Supplementary file 1 for detailed genotypes.
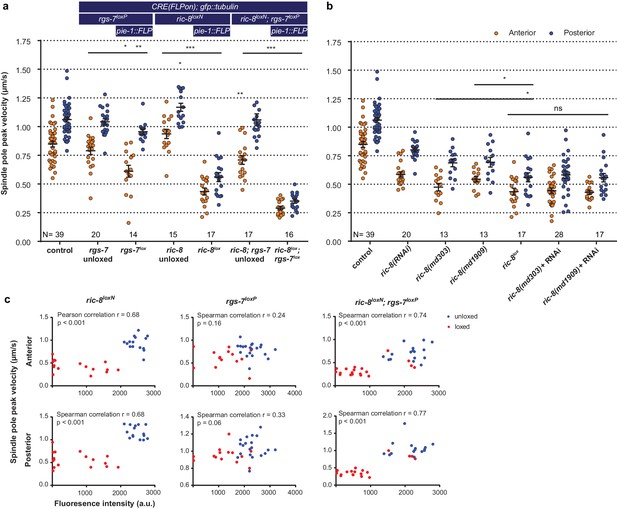
Inducible knockout of essential genes in the C. elegans germline.
(a) Spindle severing experiments of the induced rgs-7lox and ric-8lox knockout alleles compared to embryos in which FLP and the consequent recombination cascade were not induced. Control is the gfp::tubulin strain. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; *p<0.05, **p<0.01, ***p<0.001. (b) Spindle severing experiments to compare the induced ric-8lox knockout phenotype with feeding RNAi, two ric-8 partial loss of function alleles, and mutants + RNAi. Control is the gfp::tubulin strain. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; ns p<0.05, *p<0.05. (c) Scatter plots showing a correlation between loss of mcherry expression (CRE activation) and reduced spindle pole peak velocities for ric-8loxN, rgs-7loxP, and double ric-8loxN; rgs-7loxP embryos. Embryos were isolated from transgenic F1 (loxed), and control animals that were not injected with the FLP construct (unloxed) embryos. Clearly, some of the ric-8loxN; rgs-7loxP loxed cluster with the unloxed. These were discarded as outliers from further analyses. Data points were from the same experiment as in a. See Supplementary file 1 for detailed genotypes.
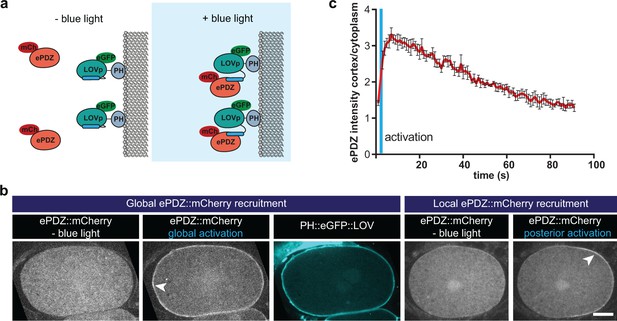
Optimized ePDZ–LOV enables light-inducible control of endogenous protein localization in the C.elegans one-cell embryo.
(a) Cartoon model illustrating the proof of concept wherein cytosolic ePDZ::mCherry is cortically recruited to membrane PH::LOV upon activation with blue light. Blue fields indicate conditions in which both ePDZ- and LOV components are present, and an ePDZ–LOV interaction is induced with blue light. (b) Spinning disk confocal images showing light-controlled localization of proteins in the C. elegans zygote (arrowheads). See Materials and methods for the local activation procedure. Also see Videos 3–5. Scale bar: 10 µm. Anterior is to the left in all microscopy images. (c) Quantification of cortical ePDZ::mCherry enrichment measured over time after a 1 s pulse activation (blue vertical line). Error bars: s.e.m. t1/2 calculated with single component non-linear regression.
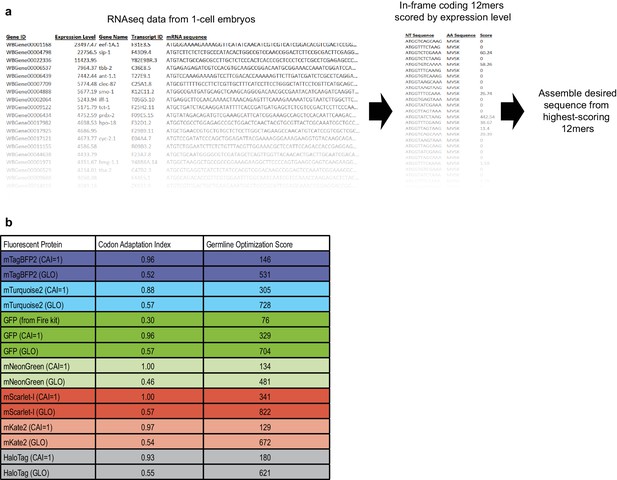
Method of germline-optimization of coding sequences confers resistance to silencing.
(a) Strategy for designing germline-optimized coding sequences. We used an RNAseq dataset from one-cell embryos (Gerstein et al., 2014; Hillier et al., 2009) as a proxy for germline-expressed mRNAs, because one-cell embryos are not transcriptionally active and thus their entire mRNA content should be derived from the maternal germline. We made a list of all 12-nucleotide coding words and assigned each word a score that is the sum of the RPKM expression values of the germline mRNAs in which that word appears. Thus, words that appear frequently, or in highly-expressed germline genes, will have high scores. Although the germline silencing machinery is presumably blind to reading frame, we only considered in-frame 12mers so that our design algorithm would implicitly account for codon usage. Finally, we developed an algorithm to assemble any desired coding sequence from our list of coding 12mers. The algorithm is as follows: (1) Based on the desired amino acid sequence, assemble a list of all 12mers that could appear in the coding sequence, and sort these words by score; (2) Assemble a draft coding sequence by plugging in one word at a time, beginning with the highest-scoring possible word and continuing until the sequence is complete; (3) Compute a score for the entire draft sequence by averaging the scores of each word it contains; (4) Refine the sequence by randomly choosing one word at a time, changing it to a different word, and checking whether the overall sequence score improves. Repeat step (4) until no further improvements are found after a certain number of iterations, which is chosen based on the sequence length such that each residue has a 99% chance of being tested at least once. The random optimization in step (4) is necessary because step (2) favors the highest-scoring words without considering context. Choosing a high-scoring word at one position constrains the subsequent choice of words that overlap the high-scoring word. In some cases, a higher overall score results from choosing two moderately-scoring words, rather than a high- and a low-scoring word, when the two words overlap. Random optimization ensures that these cases are found and maximizes the overall score of the designed sequence. (b) Results of applying our germline optimization algorithm to a selected set of fluorescent protein tags. Codon adaptation index (Redemann et al., 2011) and our germline optimization score are shown for coding sequences designed to maximize optimal codon usage (CAI = 1 indicates that sequence was designed to have a codon adaptation index as close as possible to 1.00) or to maximize the germline optimization score (GLO, GermLine Optimized). Germline optimization significantly increases the germline optimization score as expected, at the cost of a moderate reduction in codon adaptation index. Despite the lower codon adaptation index, germline-optimized transgenes were most often expressed at higher levels than their codon-optimized counterparts (see above and our unpublished observations). Constructs containing the listed germline-optimized fluorescent protein sequences will be deposited at Addgene.
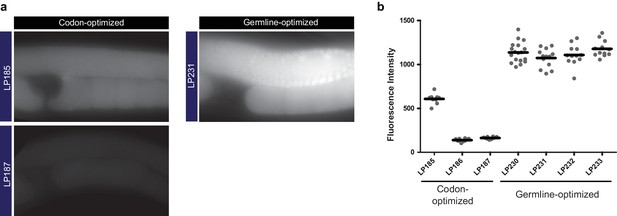
Germline-optimization of coding sequences confers resistance to silencing.
(a) Images of germline fluorescence in animals carrying either codon-optimized (Redemann et al., 2011) or germline-optimized mNeonGreen::AraD transgenes. AraD is a tetrameric bacterial protein used as a control for single-molecule fluorescence photobleacing experiments (Dickinson et al., 2017). See Supplementary file 1 for detailed genotypes. (b) Measurements of fluorescence intensity in the germlines of strains carrying either codon-optimized or germline-optimized mNeonGreen::AraD transgenes. Each data point represents one animal. The high expression of the germline-optimized transgenes was stable for >20 generations.
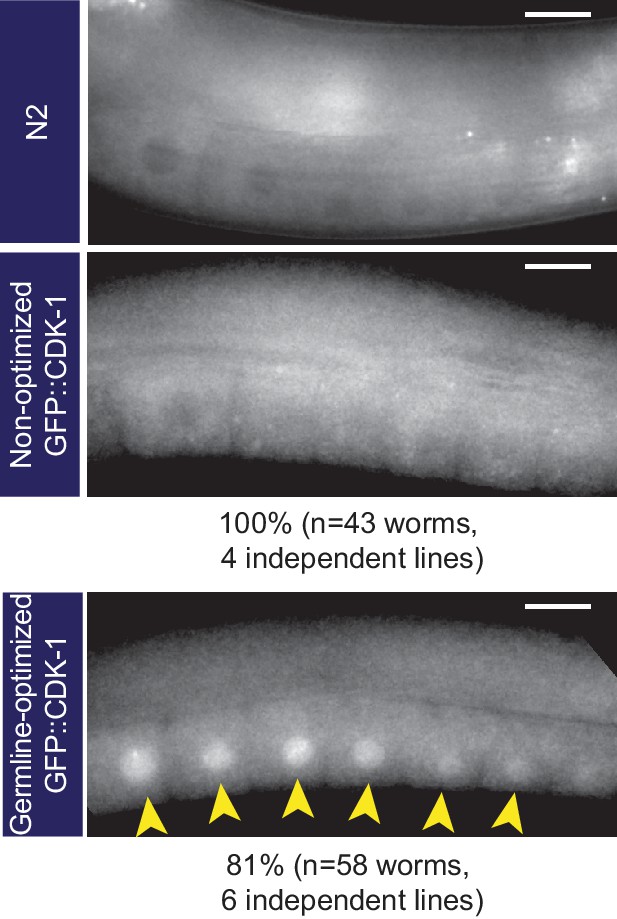
Germline-optimization can allow expression even of highly silencing-prone transgenes, although expression is transient.
Images of germline fluorescence in animals carrying GFP::CDK-1 transgenes made with either non-optimized or germline-optimized GFP. gfp::cdk-1 was tested because this transgene is especially prone to silencing (Lee et al., 2012). A non-transgenic (N2) animal is shown as an example of autofluorescence. We consistently observed expression of germline-optimized GFP::CDK-1 shortly after transgene isolation, although this expression was gradually lost over ~10 generations when we cultured these strains without selecting fluorescent animals at each passage. See Supplementary file 1 for detailed genotypes.
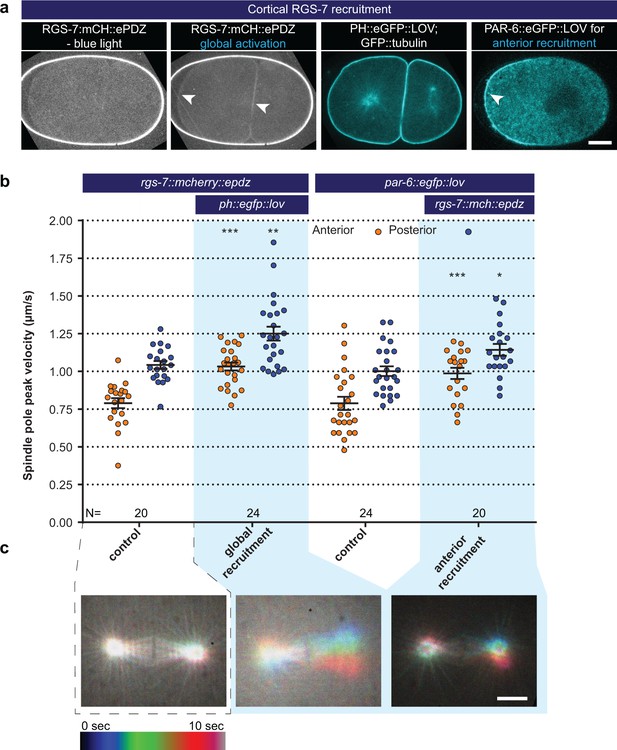
Light-controlled localization of endogenous Gα regulator RGS-7 in the C. elegans embryo.
(a) Light-controlled localization of endogenous RGS-7 to membrane PH::LOV (arrowheads, note that the eggshell shows strong autofluorescence in the red channel). Most right panel: anterior localization of PAR-6::eGFP::LOV. Scale bar: 10 µm (b) Spindle severing experiments after light-induced cortical localization of RGS-7 (blue fields). Controls are the rgs-7::mcherry::epdz and par-6::egfp::lov strains. Experimental conditions: combination with ph::egfp::lov and rgs-7::mcherry::epdz. Blue fields indicate conditions in which both ePDZ and LOV components are present, and an ePDZ–LOV interaction is induced with blue light. Blue light activation was global and continuous. Error bars: s.e.m. Welch’s Student’s t-test; *p<0.05, **p<0.01, ***p<0.001. (c) Maximum projections of spindle movements for 10 s using a temporal color coding scheme to visualize spindle movements. A stationary spindle produces a white maximum projection, whereas a mobile spindle leaves a colored trace. Scale bar: 5 µm.
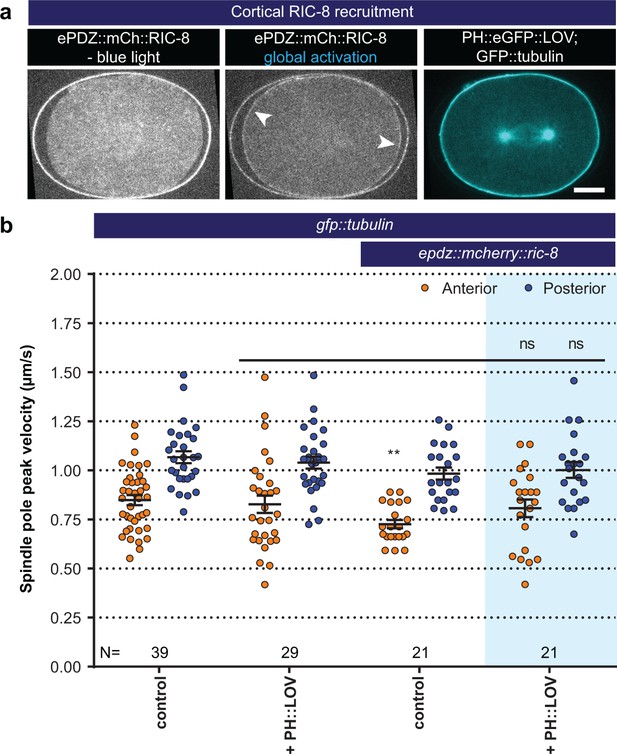
Light-controlled localization of the endogenous Gα regulator RIC-8 in the C. elegans embryo.
(a) Light-controlled localization of endogenous RIC-8 to membrane PH::eGFP::LOV (arrowheads, note that the eggshell shows strong autofluorescence in the red channel). Scale bar: 10 µm. (b) Spindle severing experiments after light-induced cortical localization of RIC-8 (blue fields). Controls are gfp::tubulin and epdz::mcherry::ric-8 strains. Experimental conditions: combination with ph::egfp::lov. Blue fields indicate conditions in which both ePDZ and LOV components are present, and an ePDZ–LOV interaction is induced with blue light. Error bars: s.e.m. Welch’s Student’s t-test; ns p>0.05. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
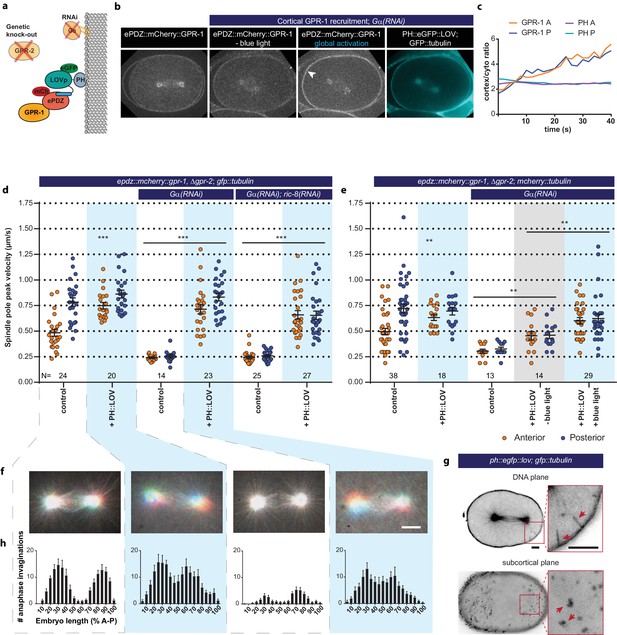
Light-inducible GPR-1 recruitment to the cortex rescues pulling force generation in the absence of Gα.
(a) Cartoon model illustrating the experiment that localizes GPR-1 directly to the membrane, bypassing the wild type membrane anchor Gα which is inactivated by RNAi. (b) Spinning disk confocal images of light-controlled cortical GPR-1 recruitment independent of the wild type anchor Gα (arrowheads; note the autofluorescent eggshell in the mCherry channel). Scale bar: 10 µm. (c) Quantification of cortical GPR-1 recruitment during continuous activation of the ePDZ–LOV interaction, represented as the ratio of cortical/cytoplasmic signal. Also see Videos 8 and 9. (d) Spindle severing experiments in combination with cortical recruitment of endogenous GPR-1. Control is the epdz::mcherry::gpr-1, Δgpr-2; gfp::tubulin strain. Experimental conditions: combinations with ph::egfp::lov, Gα(RNAi), and Gα(RNAi); ric-8(RNAi). Blue fields indicate conditions in which both ePDZ- and LOV components are present, and an ePDZ–LOV interaction is induced with blue light. Blue light activation was global and continuous. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; **p<0.01, ***p<0.001. (e) Spindle severing experiments in combination with cortical recruitment of endogenous GPR-1. Control is the epdz::mcherry::gpr-1, Δgpr-2; mcherry::tubulin strain. Experimental conditions: combinations with ph::egfp::lov, Gα(RNAi), and the absence of blue light (grey field). Blue fields indicate conditions in which an ePDZ- and LOV component are present, and an ePDZ–LOV interaction is induced using blue light. Blue light activation was global and continuous. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; **p<0.01, ***p<0.001. (f) Maximum projections of spindle movements for 10 s using a temporal color coding scheme to visualize spindle movement as in Figure 3c. (g) Plasma membrane invaginations resulting from cortical pulling forces are visible as lines in the DNA plane and dots in the subcortical plane (red arrows). Larger structures are membrane ruffles, which are distinct from the more dynamic invaginations, as can be seen in Video 11. Scale bar: 5 µm. (h) Distribution of anaphase membrane invaginations plotted along anterior-posterior embryo length. Conditions were the same as for the connected experiments in d and f, except for the control, which was the ph::egfp::lov; gfp::tubulin strain and not epdz::mcherry::gpr-1, Δgpr-2. Scale bar: 5 µm. Blue fields indicate conditions in which ePDZ and LOV components are present, and an ePDZ–LOV interaction is induced using blue light. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
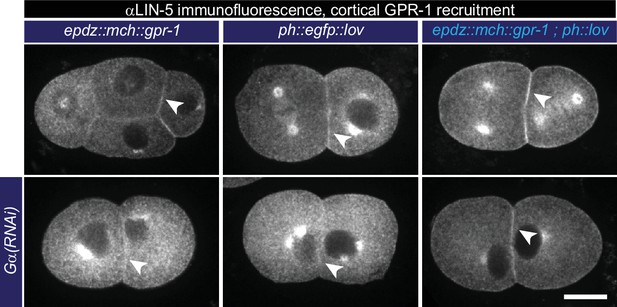
Cortical GPR-1 recruitment subsequently recruits LIN-5 to the cortex.
Immunofluorescent staining of LIN-5 in early embryos. Controls are the epdz::mcherry::gpr-1 and ph::egfp::lov strains. Experimental condition is the combination of the two controls (GPR-1 recruitment to membrane LOV) in otherwise wildtype embryos or Gα(RNAi) embryos. Arrowheads indicate cortical LIN-5. Scale bar: 10 µm. Images are maximum projections of 9 imaging planes with 0.25 µm spacing. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
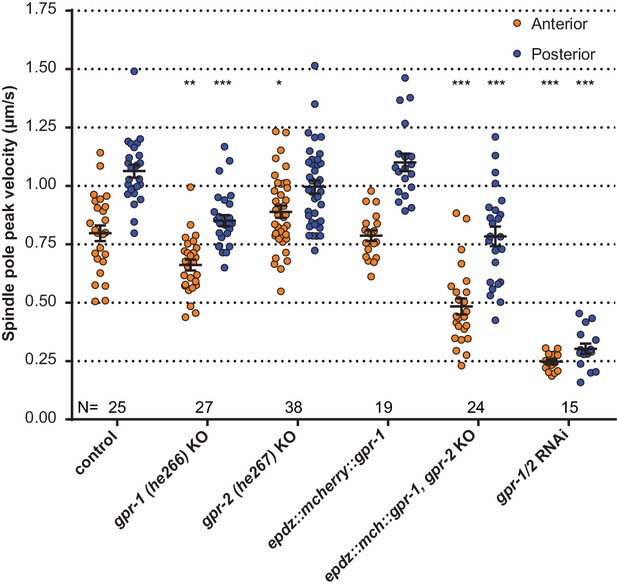
Spindle severing experiments of CRISPR/Cas9-mediated gpr-1 and gpr-2 knockout alleles, and tagged gpr-1.
egfp::tubulin (control) and gpr-1/2(RNAi) are wildtype and gpr-1/2 loss of function controls. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; *p<0.05, **p<0.01, ***p<0.001. See Supplementary file 1 for detailed genotypes.
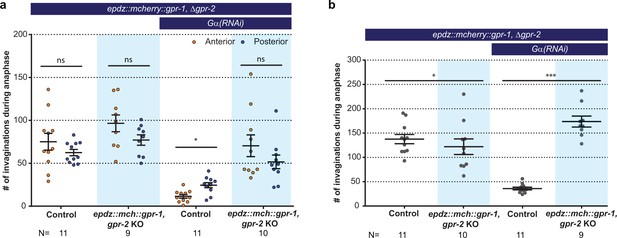
Cortical GPR-1 recruitment increases cortical pulling forces and the number of plasma membrane invaginations.
(a) Number of membrane invaginations, anterior and posterior compared for indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Paired, two-tailed Student’s t-test; ns p>0.05, *p<0.05. (b) Total number of membrane invaginations for the indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Welch’s Student’s t-test; *p<0.05, ***p<0.001. Blue fields indicate conditions in which an ePDZ- and LOV component are present, and an ePDZ–LOV interaction is induced using blue light. See Supplementary file 1 for detailed genotypes.
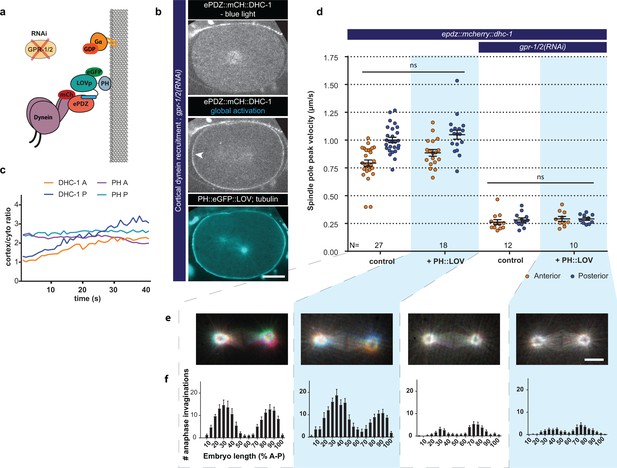
Direct cortical anchoring of dynein (DHC-1) is insufficient for cortical pulling force generation.
(a) Cartoon model illustrating the experiment where dynein is recruited directly to the cortex. The wild type force generator complex is inactivated by RNAi. (b) Spinning disk confocal images showing light-controlled recruitment of dynein to the cortex (arrowheads, note the autofluorescent eggshell in the mCherry channel). Scale bar: 10 µm. (c) Quantification of cortical dynein recruitment during continuous activation of PH::LOV with blue light. (d) Spindle severing experiments with cortical dynein recruitment. Control is the epdz::mcherry::dhc-1; gfp::tubulin strain. Experimental conditions: combination of ph::egfp::lov and gpr-1/2(RNAi). Blue light activation was global and continuous. Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; ns p>0.05. (e) Maximum projections of spindle movements for 10 s using a temporal color coding scheme to visualize spindle movement as in Figure 3c. Scale bar: 5 µm. (f) Distribution of anaphase membrane invaginations plotted along anterior-posterior embryo length. Conditions were the same as for the connected experiments in d and e, except for the control, which was the ph::egfp::lov; gfp::tubulin strain and not epdz::mcherry::dhc-1. Blue fields indicate conditions in which both ePDZ and LOV components are present, and an ePDZ–LOV interaction is induced with blue light. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
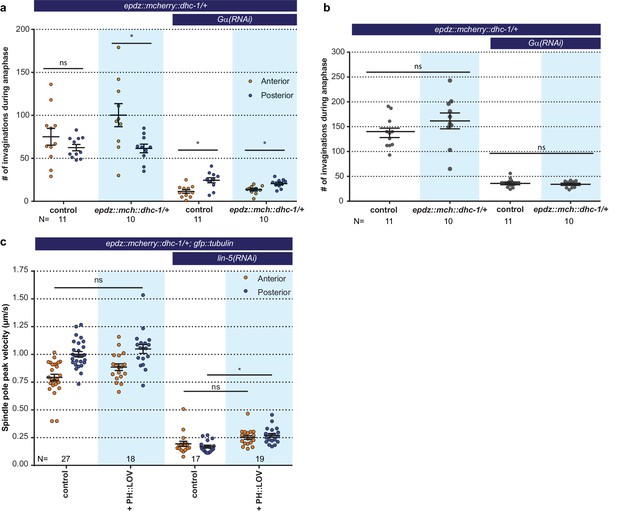
Cortical dynein (DHC-1) recruitment does not enhance cortical pulling force generation.
(a) Number of membrane invaginations, anterior and posterior compared for indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Paired, two-tailed Student’s t-test; ns p>0.05, *p<0.05. (b) Total number of membrane invaginations for the indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Welch’s Student’s t-test; ns p>0.05. (c) Spindle severing experiments with cortical dynein recruitment. Control is the epdz::mcherry::dhc-1; gfp::tubulin strain. Experimental conditions: combination of ph::egfp::lov and lin-5(RNAi). Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; ns p>0.05, *p<0.05. Blue fields indicate conditions in which an ePDZ and LOV component are present, and an ePDZ–LOV interaction is induced using blue light. See Supplementary file 1 for detailed genotypes.
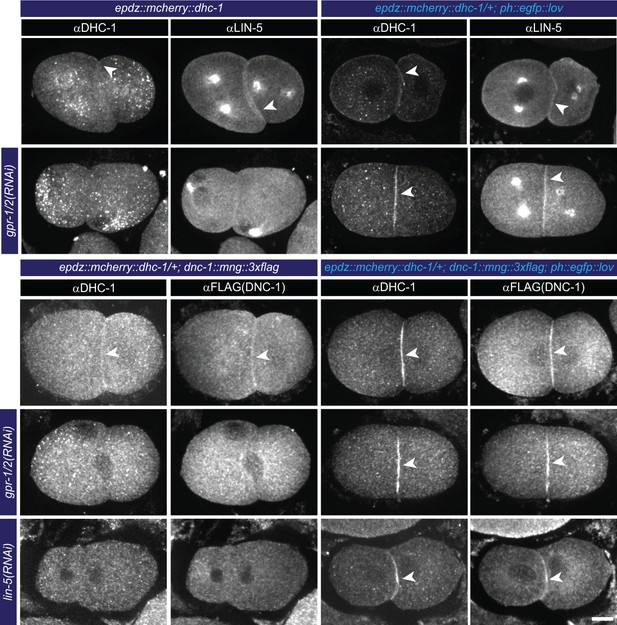
Cortical dynein (DHC-1) recruitment localizes LIN-5.
(a) Immunofluorescent staining of DHC-1, LIN-5 and DNC-1 (dynactin) in early embryos. Control is epdz::mcherry::dhc-1/+ strain. Experimental condition is cortical dynein recruitment in gpr-1/2(RNAi) or otherwise wildtype embryos. Arrowheads indicate cortical staining. Scale bar: 10 µm. Images are maximum projections of 9 imaging planes with 0.25 µm spacing. Blue light activation was global and continuous. Anterior is to the left in all microscopy images.
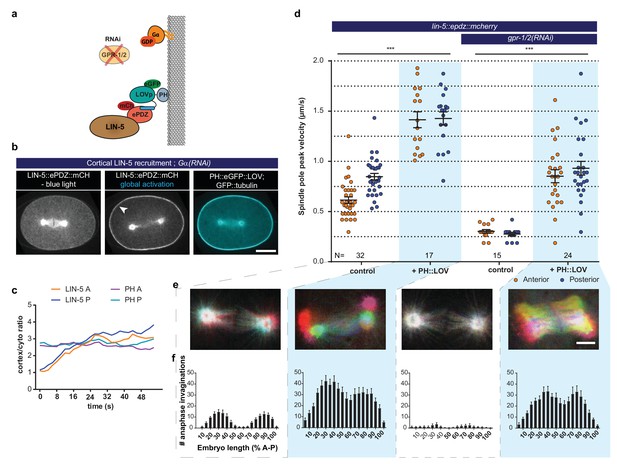
LIN-5 is a strong and essential activator of dynein-dependent cortical pulling forces.
(a) Cartoon model illustrating the experiments in which LIN-5 is recruited to the cortex independently of the wild type Gα–GPR-1/2 anchor. (b) Spinning disk confocal images showing light-controlled recruitment of endogenous LIN-5 in the absence of Gα (arrow head). See also Videos 14 and 15. (c) Cortical LIN-5 recruitment during continuous activation of the ePDZ–LOV interaction, represented as the ratio of cortical/cytoplasmic signal. Scale bar: 10 µm. (d) Spindle severing experiments in combination with cortical recruitment of endogenous LIN-5. Control is the lin-5::epdz::mcherry; gfp::tubulin strain. Experimental conditions: combinations with ph::egfp::lov and gpr-1/2(RNAi). Error bars: s.e.m. Welch’s Student’s t-test and Mann Whitney U test; ***p<0.001. (e) Maximum projections of spindle movements for 10 s using a temporal color coding scheme to visualize spindle movement as in Figure 3c. Scale bar: 5 µm. (f) Anaphase membrane invaginations plotted along anterior-posterior embryo length. Conditions were the same as for the connected experiments in d and e, except for the control, which was the ph::egfp::lov; gfp::tubulin strain and not lin-5::epdz::mcherry. Blue fields indicate conditions in which ePDZ and LOV components are present, and an ePDZ–LOV interaction is induced using blue light. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
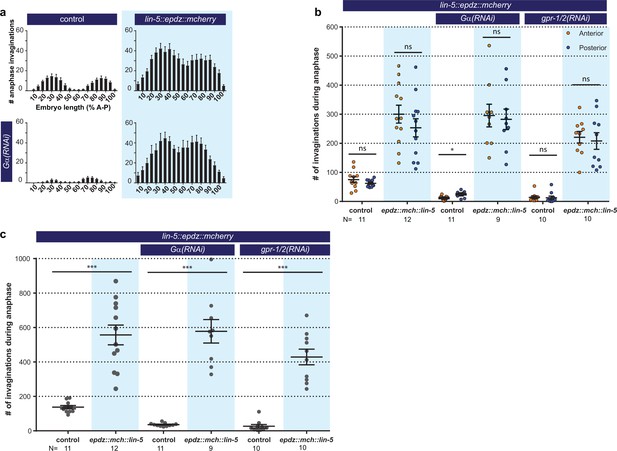
Cortical LIN-5 recruitment strongly increases cortical pulling force generation.
(a) Distribution of membrane invaginations along the one-cell embryo anterior-posterior axis. Control is the ph::egfp::lov; gfp::tubulin strain with and without Gα RNAi. Experimental condition is the combination of control with lin-5::epdz::mcherry for cortical LIN-5 recruitment. (b) Number of membrane invaginations, anterior and posterior compared for indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Paired, two-tailed Student’s t-test; ns p>0.05, *p<0.05. (c) Total number of membrane invaginations for the indicated genotypes. Control is the gfp::tubulin; ph::egfp::lov strain. Error bars: s.e.m. Welch’s Student’s t-test; ***p<0.001. Blue fields indicate conditions in which an ePDZ- and LOV component are present, and an ePDZ–LOV interaction is induced using blue light. Blue light activation was global and continuous. See Supplementary file 1 for detailed genotypes.
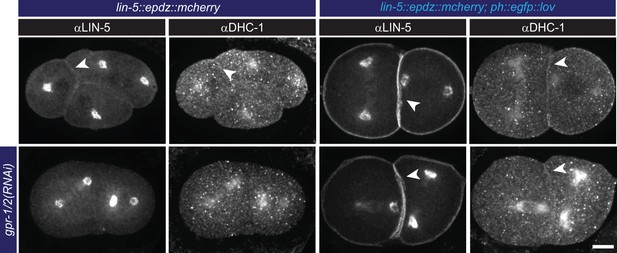
Cortical LIN-5 recruitment localizes dynein (DHC-1).
Immunofluorescent staining of LIN-5 and DHC-1 in early embryos. Control is the lin-5::epdz::mcherry strain. Experimental condition is cortical LIN-5 recruitment in gpr-1/2(RNAi) or otherwise wildtype embryos. Arrowheads indicate cortical staining. Scale bar: 10 µm. Images are maximum projections of 9 imaging planes with 0.25 µm spacing. See Supplementary file 1 for detailed genotypes. Embryos are oriented with their anterior to the left in all images.
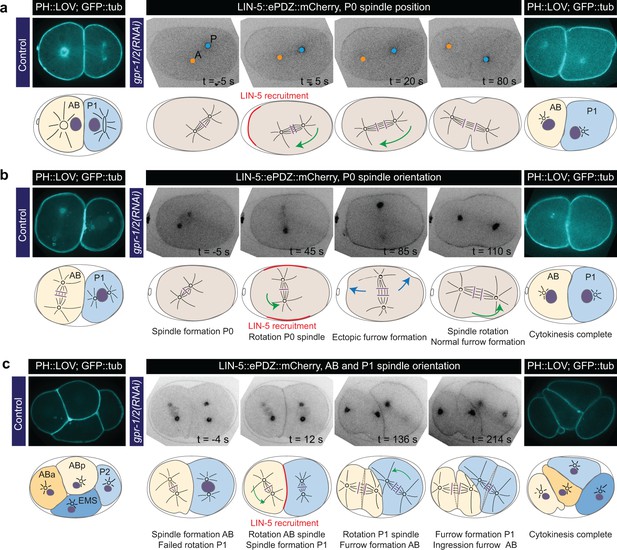
Experimentally induced spindle positioning by controlled localization of endogenous LIN-5.
(a) Selected time points of Video 18 showing induced anterior displacement of the P0 spindle upon local cortical recruitment of LIN-5. Images are annotated with centrosome positions shown as circles (orange, anterior pole; blue, posterior pole). (b) Selected time points of Video 19 showing induced transverse P0 spindle orientation upon local cortical recruitment of LIN-5. Blue arrows, ectopic furrowing. (c) Selected time points of Video 20 showing induced AB and P1 spindle rotation upon local cortical recruitment of LIN-5. In a, b, and c panels 1–4 show LIN-5::ePDZ::mCherry fluorescence, panel five shows PH::LOV and GFP::tubulin. Cartoons accompanying images illustrate key events. Red, local LIN-5 recruitment. Green arrows, spindle movements. leftmost panels show control two- and four-cell embryos labeled with PH::eGFP::LOV and GFP::tubulin. See Materials and methods for the local activation procedure. See Supplementary file 1 for detailed genotypes. Anterior is to the left in all microscopy images.
Videos
Movie montage of mitosis in a one-cell C. elegans embryo expressing GFP::tubulin (greyscale, microtubules), mCherry::TBG-1 (magenta, centrosomes) and mCherry::HIS-48 (magenta, DNA).
Images, which are single planes, were made as a time-lapse with one acquisition per 2 s and played back at 10 frames per second, with time point 0 being the final frame before the initiation of pronuclear meeting. Movie corresponds to the upper left panel in Figure 1b.
Movie montage of a mitotic spindle severing assay in a one-cell C. elegans embryo expressing GFP::tubulin (greyscale, microtubules).
The spindle is severed at the onset of anaphase using a pulsed UV laser (not visible), after which centrosomes are separated with speeds proportional to the net forces acting on them. Images, which are single planes, were made as a streaming acquisition with 0.5 s of exposure and played back at 10 frames per second, with time point 0 between late metaphase and anaphase initiation. Movie corresponds to Figure 1c.
Movie montage of a mitotic one-cell C. elegans embryo expressing diffuse cytosolic ePDZ::mCherry (greyscale, left; red, right) without PH::eGFP::LOV (green background, right).
The movie shows diffuse localization of ePDZ::mCherry in the presence of global and continuous blue light exposure, but in absence of a cortical LOV anchor. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 starting at metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to no main figure, and serves as a control for Video 4.
Movie montage of a mitotic one-cell C. elegans embryo expressing diffuse cytosolic ePDZ::mCherry (greyscale, left; red, right) and the membrane anchor PH::eGFP::LOV (green, right).
The movie shows relocalization of diffuse ePDZ::mCherry to the cortex by global and continuousactivation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 starting at late prophase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to the lower left panels in Figure 2.
Movie montage of a mitotic one-cell C. elegans embryo expressing diffuse cytosolic ePDZ::mCherry (inverted greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows relocalization of diffuse ePDZ::mCherry to the posterior cortex by local activation of cortical LOV using low-intensity blue light. Activation of the ePDZ–LOV2 interaction is induced at the posterior cortex using local illumination with a 491 nm laser. The embryo was otherwise shielded from blue light before and during the experiment. Images, which are single planes, were made as a streaming acquisition with 0.5 s of exposure and played back at 10 frames per second, with time point 0 corresponding to late prophase. Movie corresponds to the lower right panels in Figure 2.
Movie montage of a four-cell C. elegans embryo expressing diffuse cytosolic ePDZ::mCherry (greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows relocalization of diffuse ePDZ::mCherry to the cortex by activation of cortical LOV using a single pulse of blue light, and subsequent return to the dark state in the absence of blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s played back at 10 frames per second, where time point 0 is the last acquisition before a single 1 s global pulse of 491 nm light. The acquisition at time point 0 shows localization of ePDZ::mCherry in complete absence of blue light, as the embryo was kept in the dark before and after global induction of the LOV–ePDZ interaction. Movie corresponds to Figure 2c.
Movie montage of a mitotic one-cell C. elegans embryo expressing endogenously labeled ePDZ::mCherry::RIC-8 (greyscale, left; red, right), the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization ePDZ::mCherry::RIC-8 to the cortex by global and continuous activation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 starting at late prophase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry::RIC-8 in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to citation links to different figure Figure 3c.
Movie montage of a mitotic one-cell C. elegans embryo expressing endogenously labeled ePDZ::mCherry::GPR-1 (greyscale, left; red, right) in a ∆gpr-2 genetic background, the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of ePDZ::mCherry::GPR-1 to the cortex by global and continuous activation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with early metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry::GPR-1 in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie serves as a control to Video 9.
Movie montage of a mitotic one-cell C. elegans embryo treated with Gα RNAi expressing endogenously labeled ePDZ::mCherry::GPR-1 (greyscale, left; red, right) in a ∆gpr-2 genetic background, the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of ePDZ::mCherry::GPR-1 to the cortex by global and continuousactivation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with early metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry::GPR-1 in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to Figure 5b.
Movie montage of a mitotic one-cell C. elegans embryo expressing GFP::tubulin (greyscale, microtubules) and PH::eGFP::LOV (greyscale, membrane).
Invaginations (black arrows) are visible at the embryo membrane most pronouncedly in the posterior during late metaphase and anaphase. Images, which are single planes, were made as a streaming acquisition with 0.5 s of exposure and played back at 10 frames per second, with time point 0 between late metaphase and anaphase initiation. Movie corresponds to Figure 5g.
Movie montage of the subcortical area of a mitotic one-cell C. elegans embryo expressing GFP::tubulin (inverted greyscale, microtubules), PH::eGFP::LOV (inverted greyscale, membrane).
Invaginations are visible as dots protruding inwards from the embryo membrane during late metaphase and anaphase. Images, which are single planes, were made as a streaming acquisition with 0.25 s of exposure and played back at 10 frames per second, with time point 0 corresponding with anaphase, 50 s before telophase initiation. Movie corresponds to Figure 5g.
Movie montage of a mitotic one-cell C. elegans embryo expressing endogenously labeled ePDZ::mCherry::DHC-1 (greyscale, left; red, right), the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of ePDZ::mCherry::DHC-1 to the cortex by global and continuousactivation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with early metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry::GPR-1 in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie serves as a control to Video 13.
Movie montage of a mitotic one-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled ePDZ::mCherry::DHC-1 (greyscale, left; red, right), the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of ePDZ::mCherry::DHC-1 to the cortex by global and continuous activation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with early metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of ePDZ::mCherry::DHC-1 in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to Figure 6b.
Movie montage of a mitotic one-cell C. elegans embryo expressing endogenously labeled LIN-5::ePDZ::mCherry (greyscale, left; red, right) and the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of LIN-5::ePDZ::mCherry to the cortex by global and continuous activation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of LIN-5::ePDZ::mCherry in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie serves as a control to Video 15.
Movie montage of a mitotic one-cell C. elegans embryo treated with Gα RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (greyscale, left; red, right), the membrane anchor PH::eGFP::LOV and GFP::tubulin (both green, right).
The movie shows relocalization of LIN-5::ePDZ::mCherry to the cortex by global and continuous activation of cortical LOV using blue light. Images, which are single planes, were made as a time-lapse with one acquisition per 2 s for both 568 nm and 491 nm illumination and played back at 10 frames per second, with time point 0 corresponding with metaphase. The acquisition in the 568 nm channel at time point 0 shows localization of LIN-5::ePDZ::mCherry in complete absence of blue light, as embryos were kept in the dark before image acquisition. Movie corresponds to Figure 7b.
Movie montage of a mitotic one-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (not shown), the membrane anchor PH::eGFP::LOV and GFP::tubulin (both inverted greyscale).
The movie shows excessive rocking of centrosomes with associated pronuclei prior to mitotic spindle assembly. Images, which are single planes, were made as a streaming acquisition with 0.5 s of exposure and played back at 20 frames per second, with time point 0 corresponding with late prophase. Movie does not correspondto a figure, but is discussed in the text.
Movie montage of a mitotic one-cell C. elegans embryo expressing endogenously labeled LIN-5::ePDZ::mCherry (not shown) and the membrane anchor PH::eGFP::LOV (inverted greyscale).
The movie shows separation of centrosomes and associated pronuclei in prophase upon global and continuous activation of LOV with blue light. Images, which are single planes, were made as a streaming acquisition with 0.5 s of exposure and played back at 10 frames per second, with time point 0 corresponding with prophase. Movie does not correspond to a figure, but is discussed in the text.
Movie montage of a mitotic one-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (inverted greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows anterior displacement of the spindle and subsequent inverted asymmetric division resulting in a small anterior and large posterior blastomere after local recruitment of LIN-5::ePDZ::mCherry to the anterior cortex. Images, which are single planes, were made as a time-lapse with one acquisition per 5 s and played back at five frames per second, with time point 0 corresponding with metaphase. Movie corresponds to Figure 8a.
Movie montage of a mitotic one-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (inverted greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows artificial transverse positioning of the metaphase mitotic spindle and its subsequent correction to an anterior-posterior position in late anaphase after local recruitment of LIN-5::ePDZ::mCherry to the opposing equatorial cortexes. Images, which are averages of groups of 2 subsequent frames, were made as a time-lapse with one acquisition per 2 s and played back at five frames per second, with time point 0 corresponding with metaphase. Movie corresponds to Figure 8b.
Movie montage of a mitotic two-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (inverted greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows artificial rotation of transverse aligned AB and P1 spindles to an anterior-posterior position, and concurrent reorientation of the cleavage planes after local recruitment of LIN-5::ePDZ::mCherry to the central region where AB and P1 cortexes touch. Images, which are averages of groups of 2 subsequent frames, were made as a streaming acquisition with 0.5 s of exposure and played back at 10 frames per second, with time point 0 corresponding with metaphase in the AB blastomere. Movie corresponds to Figure 8c.
Movie montage of a mitotic two-cell C. elegans embryo treated with gpr-1/2 RNAi expressing endogenously labeled LIN-5::ePDZ::mCherry (inverted greyscale) and the membrane anchor PH::eGFP::LOV (not shown).
The movie shows artificial rotation of transverse aligned AB and P1 spindles to an anterior-posterior position, and concurrent reorientation of the cleavage planes after local recruitment of LIN-5::ePDZ::mCherry to the central region where AB and P1 cortexes touch. Images, which are averages of groups of 2 subsequent frames, were made as a streaming acquisition with 0.5 s of exposure and played back at 20 frames per second, with time point 0 corresponding with metaphase in the AB blastomere. Movie corresponds to Figure 8c.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| strain, strain background (Caenorhabditis elegans) | All strains derived from N2 | CGC | ||
| Antibody | mouse polyclonal anti-LIN-5 | Lorson et al., 2000 | ||
| Antibody | mouse monoclonal anti-FLAG M2 | Sigma-Aldrich | Cat. No. F1804 | |
| Antibody | rabbit polyclonal anti-DHC-1 | Gonczy et al., 1999 | ||
| software, algorithm | GLO (germline optimized) sequence optimization algorithm | this study | accessible via http://104.131.81.59/ |
Additional files
-
Supplementary file 1
Table of all C. elegans genotypes used in this study, listed per figure.
- https://doi.org/10.7554/eLife.38198.044
-
Supplementary file 2
Table of all oligonucleotides used in this study, listed per transgene.
- https://doi.org/10.7554/eLife.38198.045
-
Supplementary file 3
Table of the raw data points generated in this study, listed per figure.
- https://doi.org/10.7554/eLife.38198.046
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38198.047





