Fizzy-Related dictates A cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut
Figures
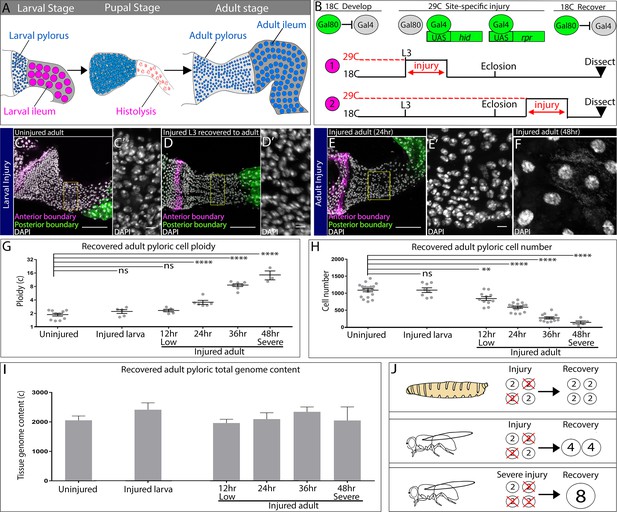
Injured hindgut pyloric cells replace lost genome content using two distinct responses.
(A) Schematic of pyloric development. (B) Experimental injury scheme (see Results and Materials and methods). Numbers 1 and 2 are referenced in the text. (C–F) Adult pylori. Anterior boundary marked by wg-LacZ (magenta), posterior boundary marked by Vha16-GFP (green), and nuclei (DAPI, white). Yellow box highlights the region shown in the adjacent high magnification inset (C’,D’,E’). (C–C’) Uninjured adult pylorus. (D–D’) Injured L3 recovered to adult (E–F). Adult pylorus injured for 24 hr (E–E’) or 48 hr (F) and recovered for 5 days. (G–H) Quantification of pyloric ploidy (G) and cell number (H). (I) Quantification of pyloric total genome content. (J) Model of larval vs adult recovery from injury. For panels (G–I) data represent mean ± SEM, N ≥ 5 animals, at least two replicates. ANOVA, Dunnett’s multiple comparisons. Scale bars (C,D,E) 50 µm, (C’,D’,E’,F’) 5 um.
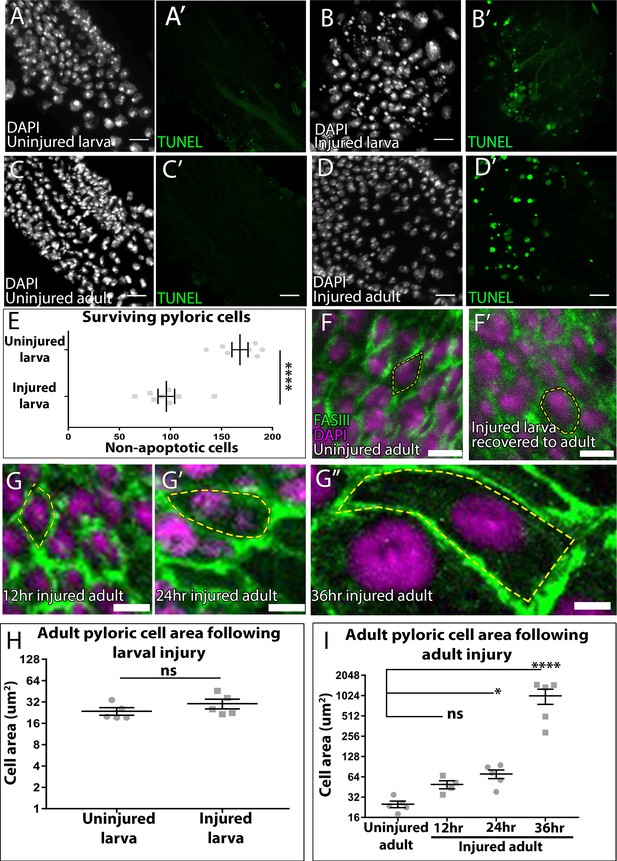
Supporting data related to Figure 1.
(A–D’) Larval and adult pylori. Cell death (TUNEL, green) and nuclei (DAPI, white). (A–A’) Uninjured larva. (B–B’) 16 hr larval injury. (C–C’) Uninjured Adult. (D–D’) 24 hr adult injury. (E) Quantification of surviving larval pyloric cells ± injury. Data represent mean ± SEM N≥5 animals, at least two replicates. Unpaired two-tailed t-test. (F–G’’’) Adult pyloric cells following injury and recovery. Membranes (FasIII, green) and nuclei (DAPI, magenta). Yellow outline highlights single cell membrane. (F) Uninjured. (F’) Larval injury. (G) 12 hr adult injury. (G’) 24 hr adult injury. (G’’) 36 hr adult injury. (H–I) Quantification of single cell area in adults ± injury as larvae (H) or adults (I). For panels (E,H,I) data represent mean ± SEM, N ≥ 4 animals, at least two replicates. Unpaired two-tailed t-test (E,H) and ANOVA with Dunnett’s multiple comparison test (I). Scale bars (A–D) 10 µm, (F–G’’) 5 µm.
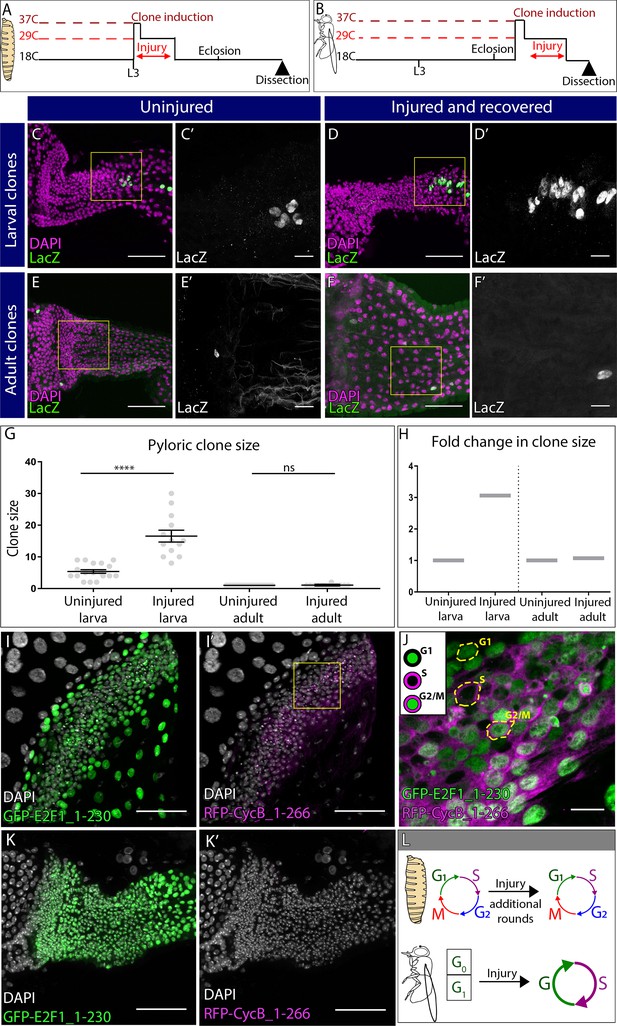
Distinct cell cycle programs underlie the distinct pyloric injury responses.
(A–B) Schematic of clone and injury induction in larvae (A) and adults (B). (C–F’) Clones in adult pylori. Clones (LacZ, green) and nuclei (DAPI, magenta). Yellow box highlights the region shown in the adjacent high magnification inset (C’,D’,E’, F’). (C–D’) Clones induced at L3 stage without injury (C–C’) or with injury (D–D’). (E–F’) Clones induced at adult stage without injury (E–E’) or with injury (F–F’). (G) Quantification of pyloric clone size ±injury. Data represent mean ± SEM, N ≥ 13 clones per condition,≥13 animals per condition, at least two replicates. Unpaired two-tailed t-tests. (H) Fold change in clone size. (I–K’) Fly-FUCCI animals expressing GFP-E2F1_1–230 (green), RFP-CycB_1–266 (magenta) and nuclei (DAPI, white). (I–I’) Fly-FUCCI in the larval pylorus. (J) Cell cycle stages in larval pyloric cells. Yellow hash marks cell outline. (K–K’) Fly-FUCCI in the adult pylorus. (L) Model of cell cycle utilized by larvae vs adults after injury. Scale bars (C,D,E,F,I,K) 50 µm, (C’,D’,E’,F’,J) =10 µm.
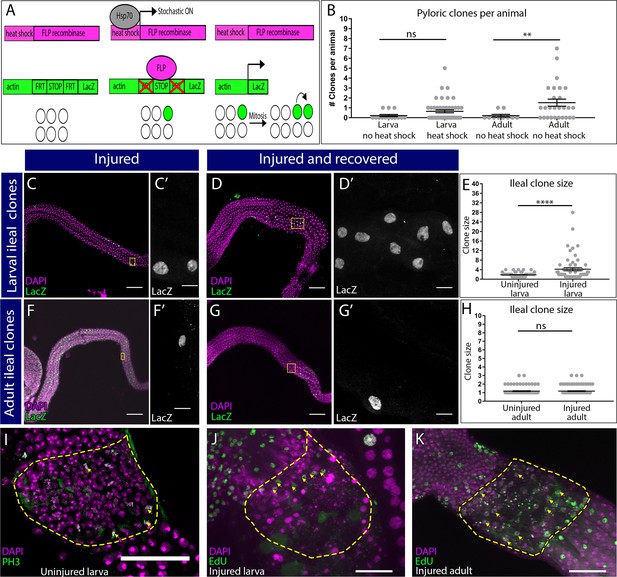
Supporting data related to Figure 2.
(A) Clonal induction system used (see Materials and methods). (B) Quantification of clones induced per animal. Data represent mean ± SEM, N ≥ 10 animals, at least two replicates. Unpaired, two-tail t-test. (C–D’) Adult ileum with clones induced at larval stage, with (D–D’) or without (C–C’) injury. Clones (LacZ, green), nuclei (DAPI, magenta). Yellow boxes highlight the region shown in adjacent high magnification insets. (E) Quantification of clone size in (C–D’). (F–G’) Adult ileum with clones induced at adult stage, with (G–G’) or without (F–F’) injury, labeling as in (C–D’). (H) Quantification of clone size in (F–G’). (I) PH3 staining (green) of Nuclei (DAPI, magenta) in the uninjured larval pylorus. (J–K) Nuclei (DAPI, magenta) and EdU incorporation (green) in injured larval (J) and injured adult (K). Yellow arrows point to EdU incorporating pyloric cells. For (E–H), data represent mean ± SEM, N ≥ 60 clones, N ≥ 10 animals, at least two replicates. Unpaired, two-tail t-test. Scale bars (C,D,F,G) 100 µm, (C’,D’,F’,G’) 10 µm, (I,J,K) 50 µm.
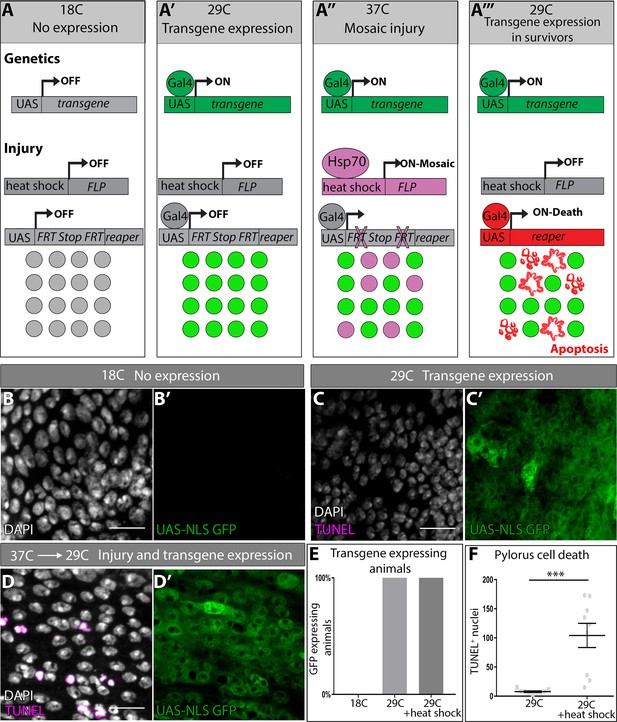
DEMISE, a novel tool for dual control of site-specific genetic ablation and transgene expression.
(A–A’’’) DEMISE method and components (see Results and Materials and methods). (B–D) Adult pylori. Transgene expression marked by UAS-NLS GFP (green), cell death (TUNEL, magenta), and nuclei (DAPI, white). (B–B’) No transgene expression at 18C. (C–C’) Transgene expression and no injury at 29C. (D–D’) Transgene expression and injury following heat shock (37C). (E) Quantification of pyloric transgene expression at different DEMISE stages. N ≥ 5 animals. (F) Quantification of pyloric cell death following heat shock. Data represent mean ± SEM, N ≥ 5 animals, at least two replicates. Unpaired two-tailed t-test. Scale bars (B–D’) 10 µm.
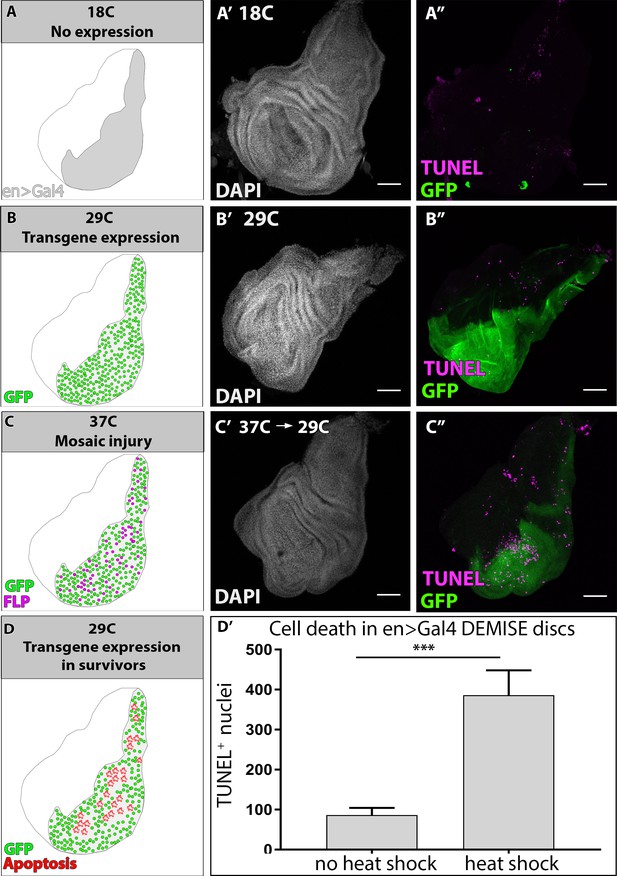
Supporting data related to Figure 3.
(A, B, C, D) Schematic of DEMISE experiment conditions for corresponding wing disc images on the right. GFP expression (green), FLP expression (magenta), apoptosis (red). (A’-A’, B’-B’, C’-C’) Nuclei (DAPI, white), transgene expression (UAS-NLS GFP, green) and TUNEL staining (magenta) of imaginal discs during DEMISE stages. (A-A’’) no transgene or site-specific cell death at low temperature (18C). (B-B’’) Transgene expression but no injury, in a site-specific manner in the engrailed region. (C-D) Heat shock activates mosaic injury within the engrailed region. (D’) Quantification of TUNEL+ nuclei within the en >Gal4 region before and after heat shock. Data represent mean ± SEM, N ≥ 5 animals, at least 2 replicates. Unpaired two-tail t-test. Scale bars 50 µm.
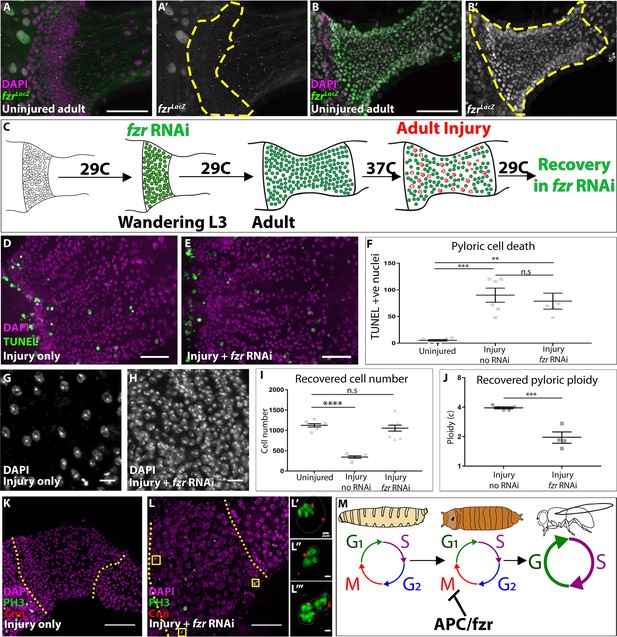
fizzy-related is a regulator of injury-mediated cell cycle programming.
(A–B’) Expression of the fzrG0418 enhancer trap (LacZ expression, green or grey), nuclei (DAPI, magenta) in uninjured larval pylorus (A–A’) or adult pylorus (B–B’). Yellow dashed line marks pyloric boundaries. (C) Schematic of DEMISE, apoptosis (red) and fzr RNAi expression (green). (D–E) Adult pylori. Cell death (TUNEL, green) and nuclei (DAPI, magenta). (D) Injury only (E) injury +fzr RNAi. (F) Quantification of pyloric cell death ±fzr RNAi. Data represent mean ± SEM, N ≥ 5 animals, at least two replicates. ANOVA with Tukey’s multiple comparisons. (G–H) Pyloric nuclei (DAPI, white) after injury and 5 days recovery. (G) injury only. (H) Injury + fzr RNAi. (I) Quantification of pyloric cell number after 5 days recovery. Data represent mean ± SEM, N ≥ 5 animals, at least two replicates per condition. ANOVA with Dunnett’s multiple comparisons. (J) Quantification of pyloric ploidy after 5 days recovery. Data represent mean ± SEM, N ≥ 4 animals, at least two replicates. Unpaired two-tailed t-test. (K–L’’’) Injured adult pylori. PH3 (green), Centrosomin (red), and nuclei (DAPI, magenta). Dashed line indicates pyloric boundaries. (K) Injury only (L) Injury + fzr RNAi. Yellow boxes highlight the region shown in adjacent high magnification insets (L’–L’’). (M) Proposed model for the role of APC/Fzr(Cdh1) in regulation of injury cell cycle responses. Scale bars (D,E) 20 µm, (G,H) 5 µm, (K,L) 50 µm (L’–L’’’) 2 µm.
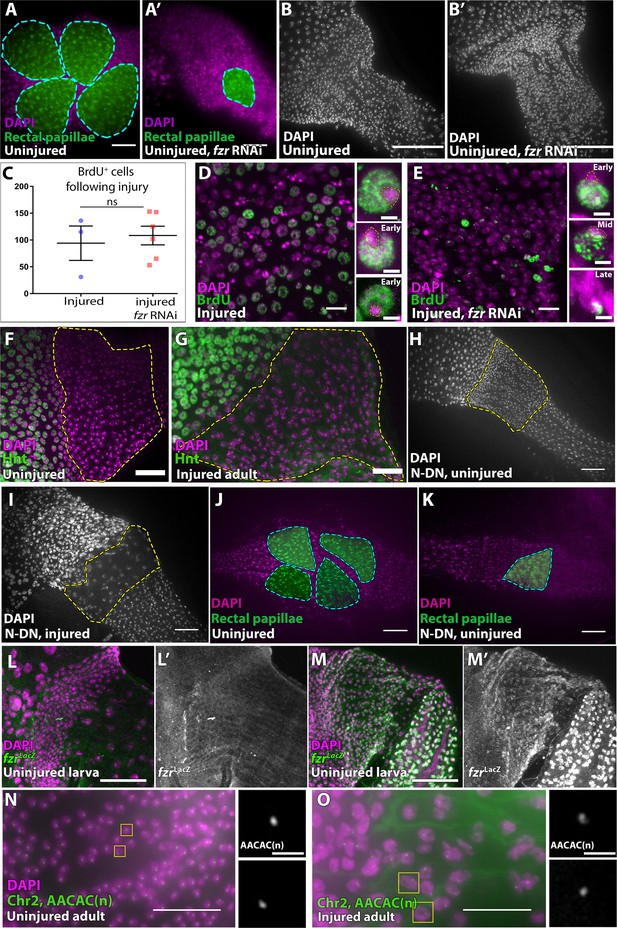
Supporting data related to Figure 4.
(A–A’) Rectal papillae, nuclei (DAPI, magenta) and Moesin-GFP (green) in control (A) and fzr RNAi (A’). (B–B’) Pyloric nuclei staining (DAPI, white) of uninjured control (B) and fzr RNAi (B’). (C) Quantification of BrdU+ cells following adult injury in the absence or presence of fzr RNAi. Data represent mean ± SEM, N ≥ 3 animals, at least two replicates. Unpaired, two-tail t-test. (D–E) Nuclei (DAPI, magenta), and EdU incorporation (green) in injured pylori of control (D) or fzr RNAi. (E). Corresponding insets highlight representative examples of EdU patterns from single cells. (F–G) The transcription factor Hindsight (green), is not expressed in pyloric nuclei (DAPI, magenta) before (F) or after (G) injury. (H–I) Pyloric nuclei (DAPI, white) of animals expressing a dominant-negative form of Notch throughout metamorphosis does not visibly alter pyloric morphology in uninjured (H) or injured animals (I). (J–K) Rectal papillae (false color, green) and nuclei (DAPI, magenta) in uninjured control(J) or Notch dominant negative animals (K) (L–M’) fzrG0326 transcription reporter (LacZ, green or grey), nuclei (DAPI, magenta) in uninjured larval pylorus (L–L’) or adult pylorus (M–M’). (N–O) DNA fluorescent in-situ hybridization for repetitive satellite region (AACAC(n), green) and nuclei (DAPI, magenta) in uninjured adults (N) and severely injured adults (O). Scale bars (A–B) 50 µm (D,E) 10 µm, insets 2 µm (F–G) 20 µm (H–K) 50 µm, (L–M’) 50 µm, (N–O) 25 µm.
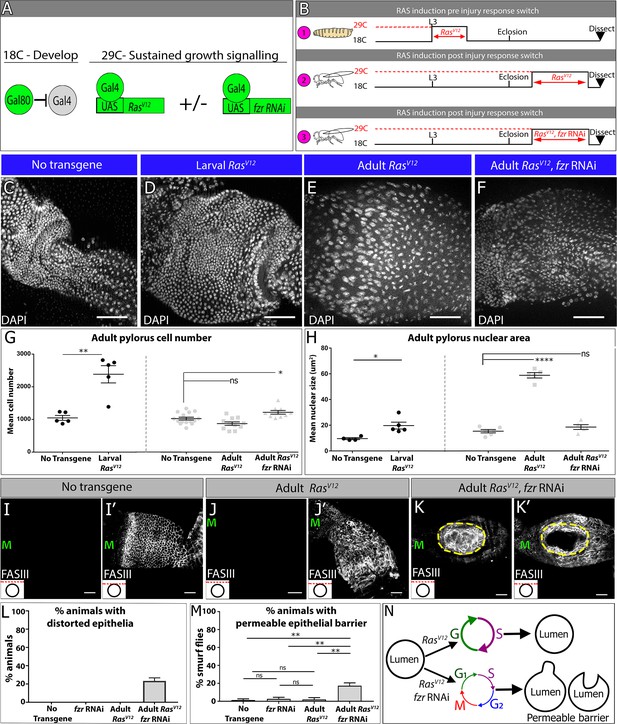
Endocycles preserve intestinal architecture in response to sustained tissue growth signaling.
(A) Schematic and (B) experimental design of sustained growth signaling expression. Numbers 1–3 in (B) are referenced in the text. (C–F) Adult pyloric nuclei (DAPI, white) ±RasVV12 expression. (C) No transgene expression. (D) 48 hr RasV12 expression at L3. (E) 6 days RasV12 expression in adults. (F) 6 days RasV12 expression in adults + fzr RNAi. (G–H) Quantification of adult pyloric cell number (G) and nuclear area (H). Data represent mean ± SEM. For adult induction, N ≥ 5 animals, at least two replicates. ANOVA with Dunnett’s multiple comparisons; For larval induction, N ≥ 4 animals, at least two replicates. Unpaired two-tailed t-test. (I–K) Adult pylorus lateral membranes highlighted by FasIII staining (white). Distinct Z-sections of the hindgut positions shown in each image are illustrated by a red line in an inset in the bottom left, where a black circle represents the hindgut epithelium. ‘M’ indicates midgut position. (I–I’) No transgene expressed. (J–J’) 14 days RasV12 expression in adults. (K–K’) 14 days RasV12 expression in adults + fzr RNAi. Yellow outline highlights distorted epithelia. (L) Quantification of % animals with epithelial distortions. Data represent mean ±SEM, N = 3 experiments, N ≥ 24 animals per condition. (M) Quantification of % animals with permeable epithelial barrier. Data represent mean ± SEM, N = 3 experiments, N ≥ 27 animals per condition. ANOVA with Tukey’s multiple comparisons. (N) Model of tissue architecture in conditions of sustained growth signaling. Scale bars (C–F,I–K’) 50 µm.
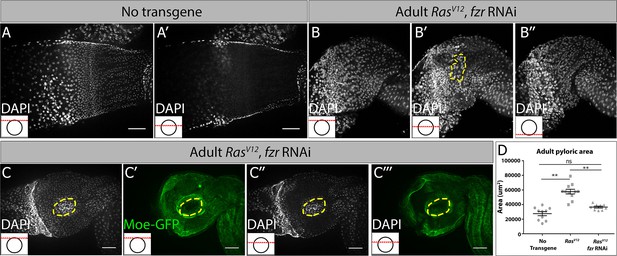
Supporting data related to Figure 5.
(A,B) Adult pylori, nuclei (DAPI, white). Distinct Z-sections of the hindgut positions shown in each image are illustrated by a red line in an inset in the bottom left, where a black circle represents the hindgut epithelium. (A,A’) No transgene control, with most epithelial cells found in a single plane. (B-B’) RasV12, fzr RNAi, with numerous epithelial cells in multiple Z-planes, highlighting tissue architecture distortion. (C-C’’’) RasV12, fzr RNAi nuclei (DAPI, white, C and C’’) and cell membranes (green, C’ and C’’’) of RasV12, fzr RNAi animals. Yellow hashed highlights large regions of distorted tissue architecture. (D) Quantification of pyloric surface area in adults expressing no transgene, RasV12 or RasV12 and fzr RNAi. Data represent mean ± SEM. N ≥ 5 animals, at least two replicates. ANOVA with Tukey’s multiple comparisons.
Tables
DEMISE combinations tested in the hindgut
https://doi.org/10.7554/eLife.38327.009| Flip-out reaper insertion | Genotype | [ry[+t7.2]=hsFLP]1 | [ry[+t7.2]=hsFLP]12 |
|---|---|---|---|
| UAS-rprFLPout 10–3 | pUAST-FRT-STOP-FRT-rpr/CyO #Insertion 10–3 | Leaky | Not leaky (used in paper) |
| UAS-rprFLPout 9–1 | pUAST-FRT-STOP-FRT-rpr/TM3Sb #Insertion 9–1 | Leaky | Not leaky |
| UAS-rprFLPout 5–1 | pUAST-FRT-STOP-FRT-rpr/TM3Sb #Insertion 5–1 | Leaky | |
| UAS-rprFLPout 4–1 | pUAST-FRT-STOP-FRT-rpr/TM3Sb #Insertion 4–1 | Not leaky | |
| UAS-rprFLPout 10–1 | pUAST-FRT-STOP-FRT-rpr/CyO #Insertion 10–1 | Leaky | Leaky |
| UAS-rprFLPout 10–2 | pUAST-FRT-STOP-FRT-rpr/TM3Sb #Insertion 10–2 | Leaky | Leaky |
| Reagent type (species) | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| gene (D. melanogaster) | fzr | NA | FBgn0262699 | |
| genetic reagent (D. melanogaster) | byn > Gal4 | Singer et al.,1996 | FBal0137290 | P{GawB}bynGal4 |
| genetic reagent (D. melanogaster) | act >> LacZ | BDSC | Stock 6355 | P{ry[+t7.2]=Act5C (FRT.polyA)lacZ.nls1}3, ry[506] |
| genetic reagent (D. melanogaster) | hsFLP12;Sco/CyO | BDSC | Stock 1929 | P{ry[+t7.2]=hsFLP} 12, y[1] w[*]; sna[Sco]/CyO |
| genetic reagent (D. melanogaster) | UAS fzr RNAi | VDRC | Stock 25550 | w1118; P{GD9960}v25550 |
| genetic reagent (D. melanogaster) | UAS-Ras85DV12 | BDSC | Stock 4847 | w[1118]; P{w[+mC]=UAS-Ras85D.V12}TL1 |
| genetic reagent (D. melanogaster) | UAS-hid, UAS-reaper | Zhou et al 1997 | ||
| antibody | anti-Fasciclin III | DSHB | 7G10 | |
| antibody | anti-Phospho-Histone H3 | Cell Signaling | ab9706 | |
| antibody | anti-BrdU | Serotec | 3J9 | |
| antibody | anti-Beta-Galactosidase | Abcam | ab9361 | |
| recombinant DNA reagent | pUAST-FRT-Stop -FRT-rpr (DEMISE) plasmid | this paper | progenitor: pUAST-FRT-Stop-FRT-mCD8-GFP | |
| recombinant DNA reagent | pUAST-FRT-Stop- FRT-mCD8-GFP plasmid | Potter et al., 2010 | addgene #24385 | |
| commercial assay or kit | TUNEL | Roche | #12156792910 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38327.014





