The decoration of specialized metabolites influences stylar development
Figures
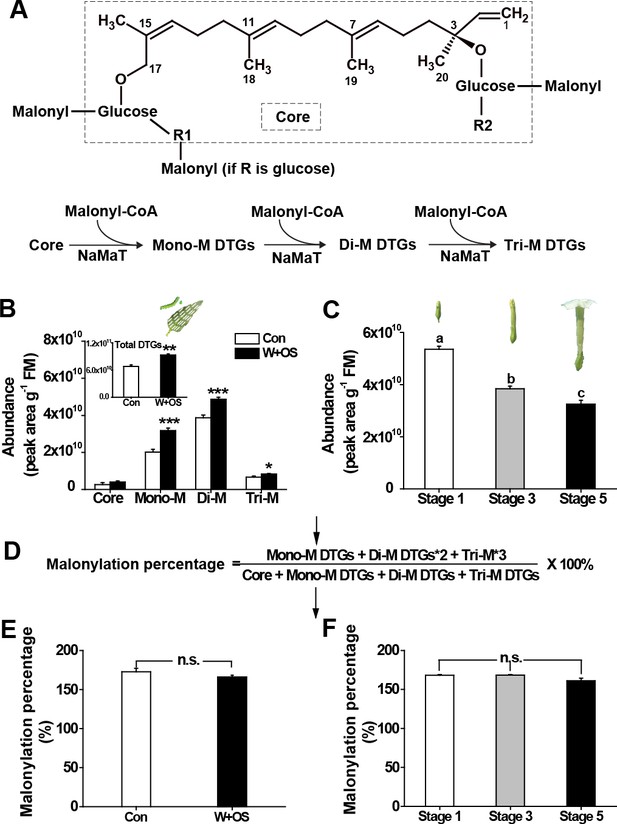
The malonylation percentage of DTGs is highly uniform across treatments and developmental stages.
(A) Structures of 17-DTGs and their malonylation options. R1 could be glucose, rhamnose or hydrogen. R2 could be rhamnose or hydrogen. NaMaT indicates malonyltransferase in N. attenuata. (B) Relative abundance (mean + SE; n = 5 – 6) of different malonylated DTGs in N. attenuata leaves, after herbivory elicitation by immediately applying M. sexta oral secretions to freshly created leaf puncture wounds (W + OS), in comparison to the same nodal positions of plants without any treatment (Con). Inset: Total DTG abundance in control and W + OS elicited leaves. (C) Total DTGs in different stages of floral development. The formula (D) describes how malonylation percentage was calculated. (E, F) Malonylation percentage (mean + SE; n = 5 – 6) of DTGs in leaves treated by W + OS (D) or different developmental flower stages (E). Asterisks indicate significant differences between controls and treatments (*p<0.05; ***p<0.001; Student’s t-tests). Different letters indicate significant differences among floral developmental stages (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
-
Figure 1—source data 1
DTG profiles and malonylation percentages under different treatments and developmental stages.
- https://doi.org/10.7554/eLife.38611.006
-
Figure 1—source data 2
DTG profiles and malonylation percentages across different tissues.
- https://doi.org/10.7554/eLife.38611.007
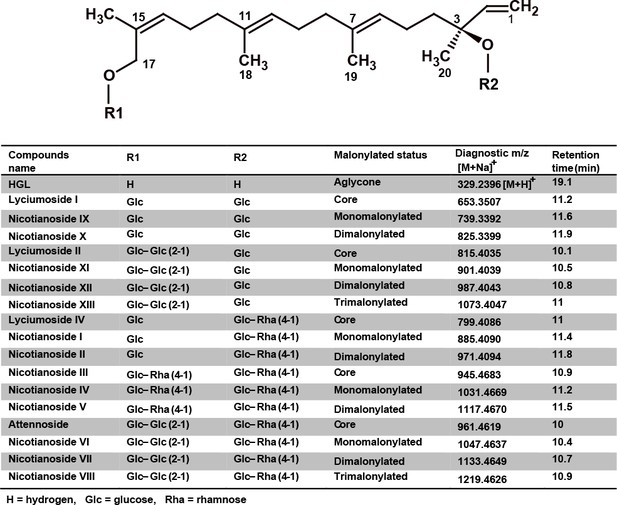
Structural diversity of N. attenuata DTGs, their diagnostic molecular ion [M + Na]+ and retention times.
https://doi.org/10.7554/eLife.38611.004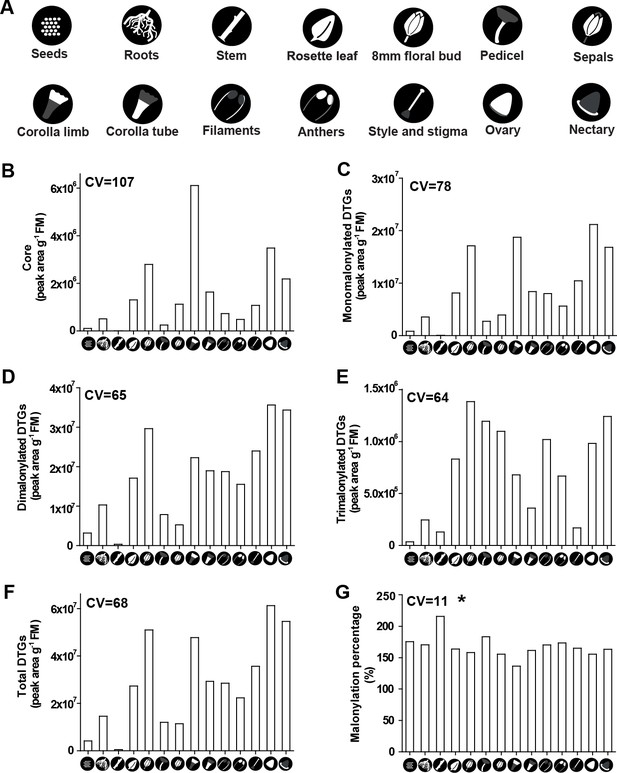
The DTG malonylation percentage is remarkably uniform across different tissues.
Fourteen tissues were collected from 28- and 50-d-old N. attenuata plants for DTGs analysis. Different tissues are represented by the symbols in (A). Relative abundance of different malonylated DTGs, including core (B), monomalonylated (C), dimalonylated (D), trimalonylated (E), and total DTGs (F). Malonylation percentage (G) was calculated based on DTG abundance. CV in each histogram represents the coefficient of variation of different malonylated DTGs and malonylation percentage. Asterisk indicates CV of (G) is an outlier based on the 3X interquartile range.
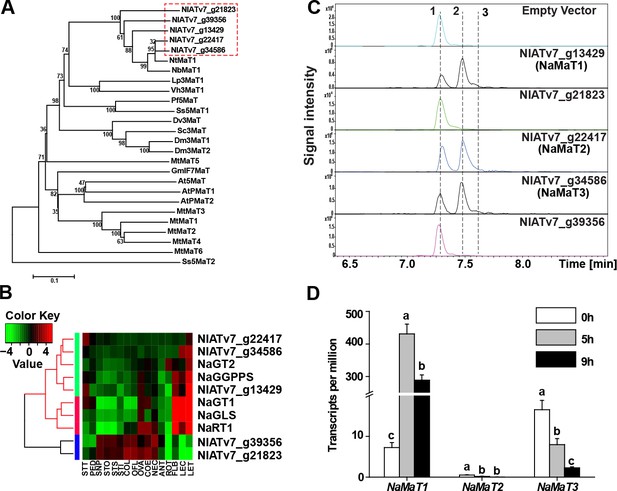
NaMaT1 mediates DTG malonylation in N. attenuata.
(A) Phylogenetic analysis of potential N. attenuata malonyltransferases (genes in red dotted box) and functionally characterized malonyltransferases in other species by amino acid sequence with accession number shown in Supplementary file 1. (B) Heatmap representing the expression of malonyltransferases and reported DTG biosynthetic genes in N. attenuata. LET, leaf treated (25 hr after wounding and elicitation with M. sexta oral secretions); LEC, leaf control; STT, stem treated; PED, pedicels; SNP, style without pollination; STO, style outcrossed; STS, style selfed; STI, stigma; COL, corolla late; OFL, opening flower; OVA, ovary; COE, corolla early; NEC, nectaries; ANT, anthers; ROT, root OS-elicited; FLB, flower bud. (C) Extracted ion chromatograms of m/z 271.2420, corresponding to the DTG aglycone, of in vitro assay products of recombinant malonyltransferases. Peaks 1, 2 and 3 were identified as Lyciumoside IV, Nicotianoside I and Nicotianoside II, respectively. (D) NaMaT1, NaMaT2 and NaMaT3 transcript counts (mean + SE; n = 3) were analyzed from RNAseq data of M. sexta-attacked leaves at indicated time points. Different letters indicate significant differences among treated time points (p<0.05, one ANOVA followed by Tukey’s HSD post-hoc tests).
-
Figure 2—source data 1
Relative transcript abundance of DTG biosynthesis genes in N. attenuata.
- https://doi.org/10.7554/eLife.38611.010
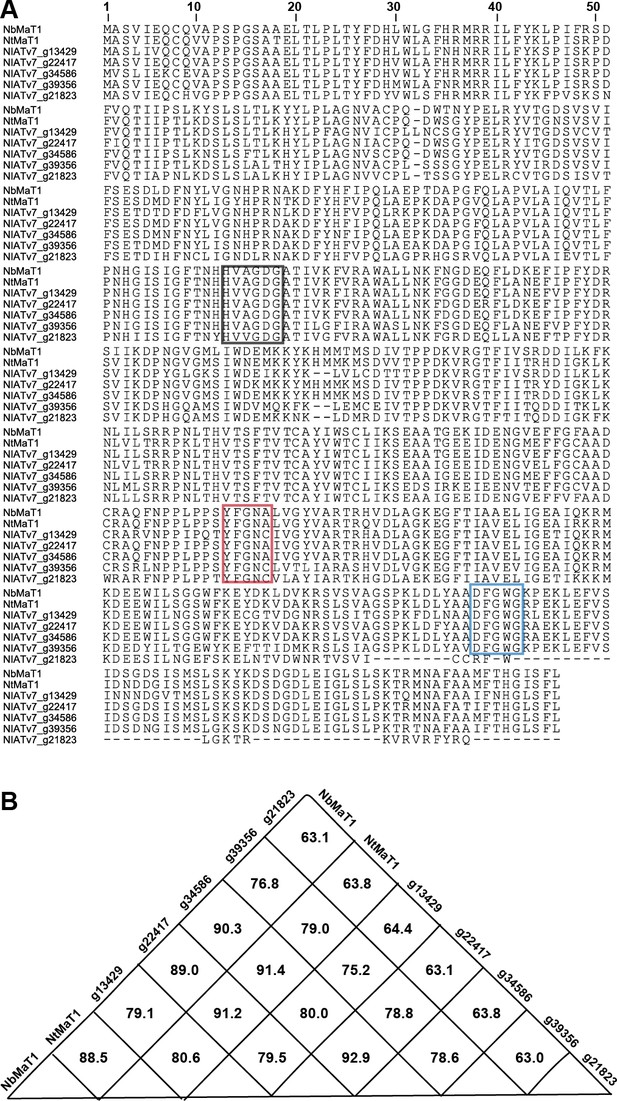
Analyzing protein sequences of NbMaT1, NtMaT1 and malonyltransferases in N. attenuata.
(A) The two conserved motifs of BAHD family members: HXXXDG (black rectangle), located near the protein center; and DFGWG (blue rectangle), located near the carboxyl terminus. Also shown is the flavonoid BAHD acyltransferase diagnostic motif YFGNC (red rectangle), which is not fully conserved in all BAHD members. (B) Protein similarity of these malonyltransferases.
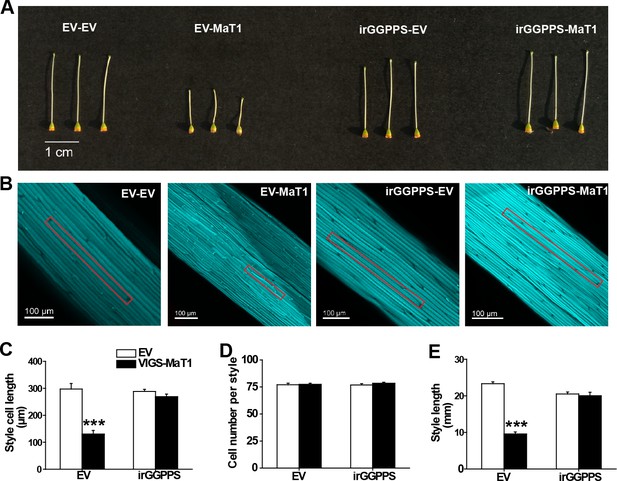
Silencing NaMaT1 expression dramatically shortens style lengths by decreasing cell size rather than cell number in a GGPPS-dependent manner.
VIGS of NaMaT1 (VIGS-MaT1) versus a control VIGS empty vector (VIGS-EV) were infiltrated in EV and irGGPPS stably-transformed plants, referred to respectively as EV-EV, EV-MaT1, irGGPPS-EV and irGGPPS-MaT1. (A) Representative photographs of gynoecia from opened flowers are shown from the indicated genotypes. (B) Typical aniline blue stained style samples from indicated genotypes. Representative cells are highlighted with a vermilion rectangle. Mean (+ SE; n = 5 – 7) cell length (C) and cell number per style (D) of styles of opened flowers of indicated plants were measured with fluorescence microscopy. (E) Style lengths (+ SE; n = 5) were measured from flowers on the first day of opening. Asterisks indicate significant differences between VIGS-EV and VIGS-MaT1 (***p<0.001; Student’s t-test).
-
Figure 3—source data 1
Silencing NaMaT1 expression dramatically shortens style lengths by decreasing cell size rather than cell number in a GGPPS-dependent manner.
- https://doi.org/10.7554/eLife.38611.015
-
Figure 3—source data 2
Effect of silencing NaMaT1 on gene expression in leaves.
- https://doi.org/10.7554/eLife.38611.016
-
Figure 3—source data 3
VIGS of NaMaT1 affects the fertility of N. attenuata plants.
- https://doi.org/10.7554/eLife.38611.017
-
Figure 3—source data 4
VIGS of NaMaT1 in both irGGPPS and irAOC backgrounds did not affect NaYUC-like 2 transcript accumulation in styles.
- https://doi.org/10.7554/eLife.38611.018
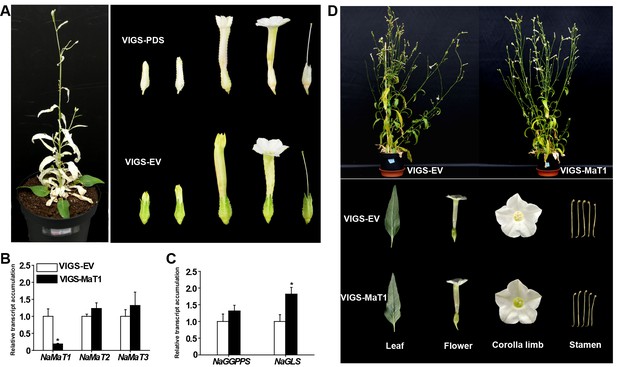
Effect of silencing NaMaT1 on gene expression in leaves, and plant morphology.
(A) Representative picture of a N. attenuata plant in which the phytoene desaturase (NaPDS) gene was silenced using VIGS (left panel) and comparison of flowers between VIGS-PDS and VIGS-EV (right panel). (B) Relative transcript levels (mean + SE; n = 7) of NaMaT1, NaMaT2, NaMaT3 and (C) NaGGPPS, NaGLS in VIGS-EV and VIGS-MaT1 leaves, 3 days after treatment with MeJA. Relative transcripts measured by RT-qPCR. (D) Leaves, flowers, corolla limbs, stamens and overall growth form of 75-day-old plants are compared to show the morphological phenotypes of entire plants (top panel) and representative parts (bottom panel). Asterisks indicate significant differences between VIGS-EV and VIGS-MaT1 (*p<0.05; Student’s t-test).
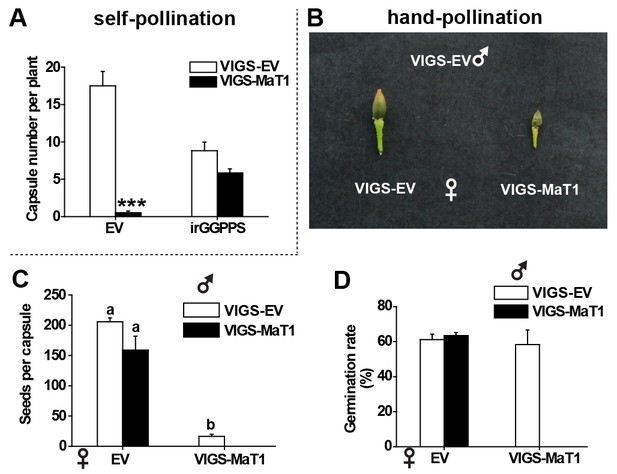
VIGS of NaMaT1 affects the fertility of N. attenuata plants.
(A) Capsule numbers (mean + SE; n = 6) of self-pollinated early senescing plants (80 days old) of the indicated genotypes. (B) The appearance of hand-pollinated capsules. Pollen from VIGS-MaT1 and VIGS-EV flowers were used to hand-pollinate emasculated flowers of VIGS-MaT1 and VIGS-EV plants. Seeds from hand-pollinated capsules were evaluated and then used for a germination test. (C) Seed numbers (mean + SE; n = 3 – 4) of hand-pollinated capsules from the indicated male and female genotypes. (D) Germination rates (mean + SE; n = 3 – 4) of seeds from hand-pollinated capsules. Asterisks indicate significant differences between VIGS-EV and VIGS-MaT1 (***p<0.001; Student’s t-test). Different letters indicate significant differences among different types of hand-pollination (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
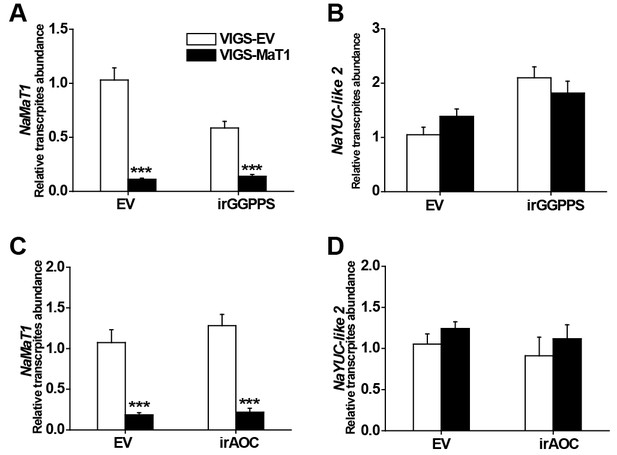
VIGS of NaMaT1 in both irGGPPS and irAOC backgrounds did not affect NaYUC-like 2 transcript accumulation in styles.
VIGS was conducted in the background of irGGPPS, irAOC and EV plants. Relative transcript abundance (mean + SE; n = 7 – 8) of NaMaT1 and NaYUC-like 2 was analyzed by qRT-PCR in styles dissected from flowers one day before anthesis. Asterisks indicate significant differences between VIGS-EV and VIGS-MaT1 plants (***p<0.001; Student’s t-test).
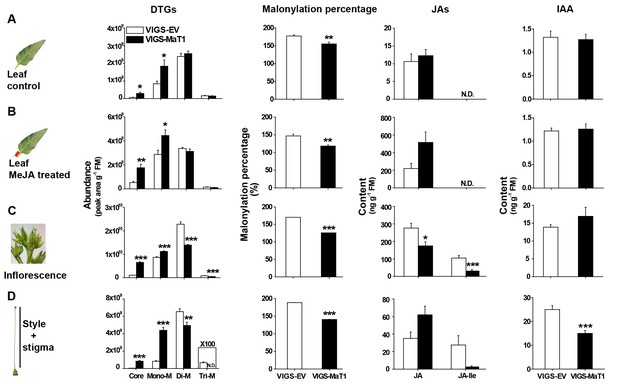
Silencing NaMaT1 influences DTG profiles and malonylation percentages across all tissues, but alters JAs and IAA specifically in flowers.
S1 leaves from early elongated VIGS plants were treated with 20 µL lanolin paste (A, leaf control) or 150 µg MeJA in lanolin paste (B, leaf MeJA treated) and samples were harvested after 3 days. The inflorescence (C), style and stigma (D) were harvested from flowering plants. Relative abundance of different malonylated DTGs (first column) were analyzed from upper samples. Malonylation percentage (second column) was calculated based on the DTG data using the formula (Figure 1). Jasmonates (JAs, third column) and IAA (fourth column) were analyzed from the same samples as those used for DTG quantification. N.D. indicates compounds which were not detected because of low concentrations. Asterisks above each column indicate significant differences between EV and VIGS-MaT1 plants (*p<0.05; **p<0.01; ***p<0.001; Student’s t-test).
-
Figure 4—source data 1
Silencing NaMaT1 influences DTG profiles and malonylation percentages across all tissues, but alters JAs and IAA specifically in flowers.
- https://doi.org/10.7554/eLife.38611.022
-
Figure 4—source data 2
DTG profiles in VIGS plants having irGGPPS, irAOC, or EV backgrounds.
- https://doi.org/10.7554/eLife.38611.023
-
Figure 4—source data 3
IAA and DTGs accumulate in high concentrations in the gynoecium.
- https://doi.org/10.7554/eLife.38611.024
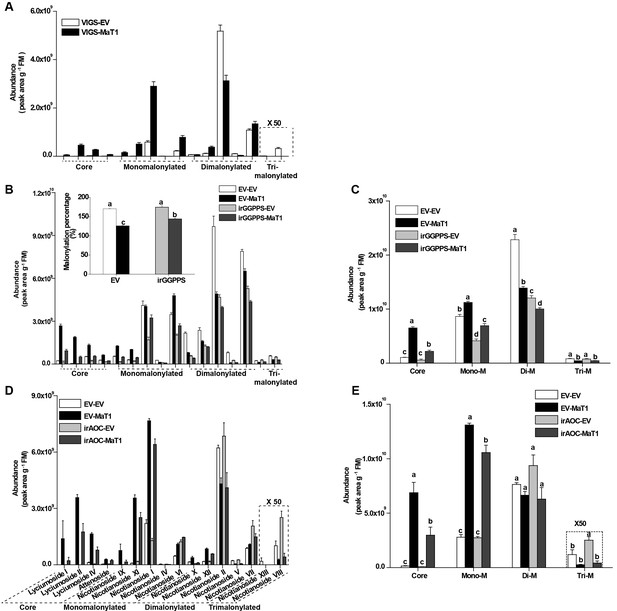
DTG profiles in VIGS plants having irGGPPS, irAOC, or EV backgrounds.
(A) Individual DTGs abundance (mean + SE; n = 9) in the styles of VIGS-MaT1 and VIGS-EV in the WT background. (B) Individual DTGs abundance (mean + SE; n = 9) in the inflorescences of VIGS-MaT1 and VIGS-EV in the background of irGGPPS and EV plants. Insert: malonylation percentages (mean + SE; n = 9) of DTGs. (C) DTGs with the same number of malonyl moieties were summed using data from (B). (D) Individual DTGs abundance (mean + SE; n = 3) in the styles of VIGS-MaT1 and VIGS-EV in background of irAOC and EV plants. (E) DTGs with the same number of malonyl moieties were summed using data from (D). Different letters indicate significant differences between genotypes (p<0.05, one-way ANOVA by Tukey’s HSD post-hoc tests). The statistic results for individual compounds are provided in the source data files.
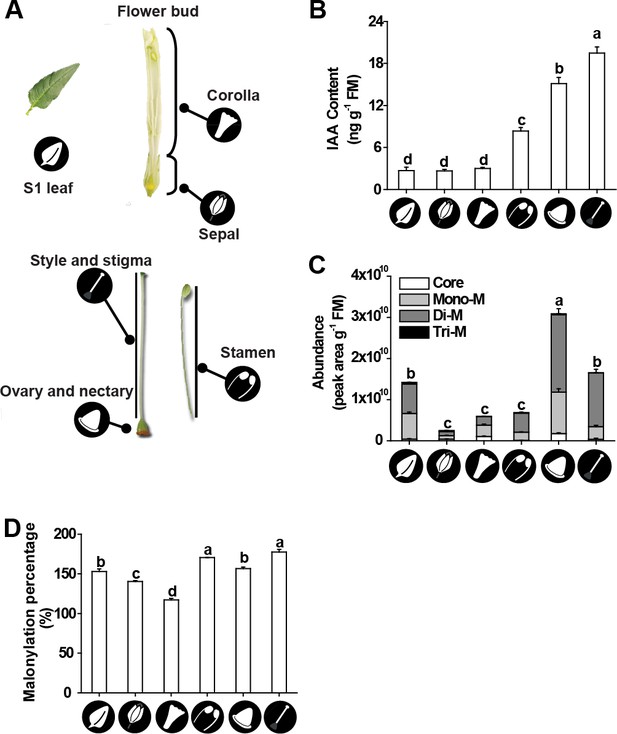
IAA and DTGs accumulate in high concentrations in the gynoecium.
S1 leaves and flower buds (one day before anthesis) were collected from flowering plants. (A) Tissues are represented by symbols as indicated. (B) IAA levels (mean + SE; n = 5) were analyzed from indicated tissues. DTG profiles (C) and their malonylation percentages (D) were analyzed using the same samples. Different letters indicate significant differences between different tissues (p<0.05, one-way ANOVA by Tukey’s HSD post-hoc tests).
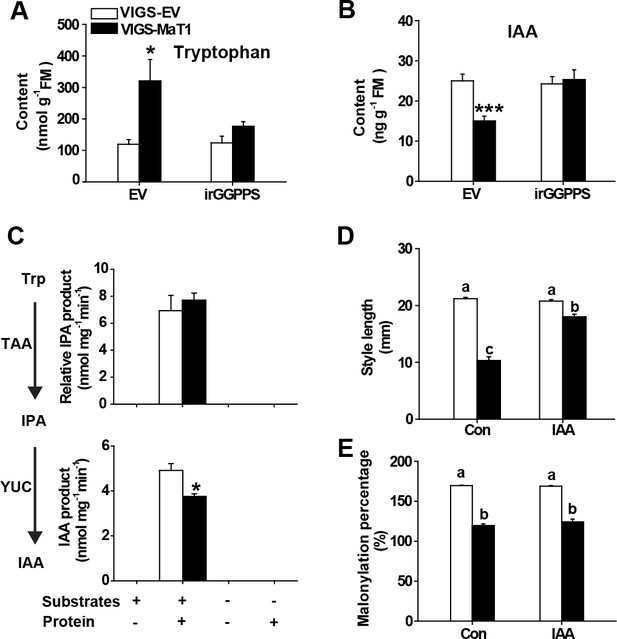
The short styles of VIGS-MaT1 flowers are associated with reduced IAA biosynthesis.
Tryptophan (A) and IAA (B) levels (mean + SE; n = 6) were analyzed from style samples dissected from flowers one day before anthesis. Crude protein was extracted from the same set of samples and then used for in vitro IAA biosynthetic enzyme activity assay. Relative in vitro enzyme assay product abundance (mean + SE; n = 5) of IPA (C, top panel) and IAA (C, bottom panel), respectively. (D) Style length (mean + SE; n = 10) of flowers injected with 0.1% DMSO (Con) or 0.1% DMSO together with 10 µM IAA 2 days before anthesis. (E) Malonylation percentage (mean + SE; n = 6) was calculated from DTGs from the same set of samples. Asterisks indicate significant differences between VIGS-EV and VIGS-MaT1 plants (*p<0.05; ***p<0.001; Student’s t-test). Different letters indicate significant differences among different lines or treatments (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
-
Figure 5—source data 1
The short styles of VIGS-MaT1 flowers are associated with reduced IAA biosynthesis.
- https://doi.org/10.7554/eLife.38611.028
-
Figure 5—source data 2
Relative abundance of flavonoids in NaMaT1 and/or NaGGPPS silenced plants.
- https://doi.org/10.7554/eLife.38611.029
-
Figure 5—source data 3
Brassinosteroid does not contribute to the VIGS-MaT1 short style.
- https://doi.org/10.7554/eLife.38611.030
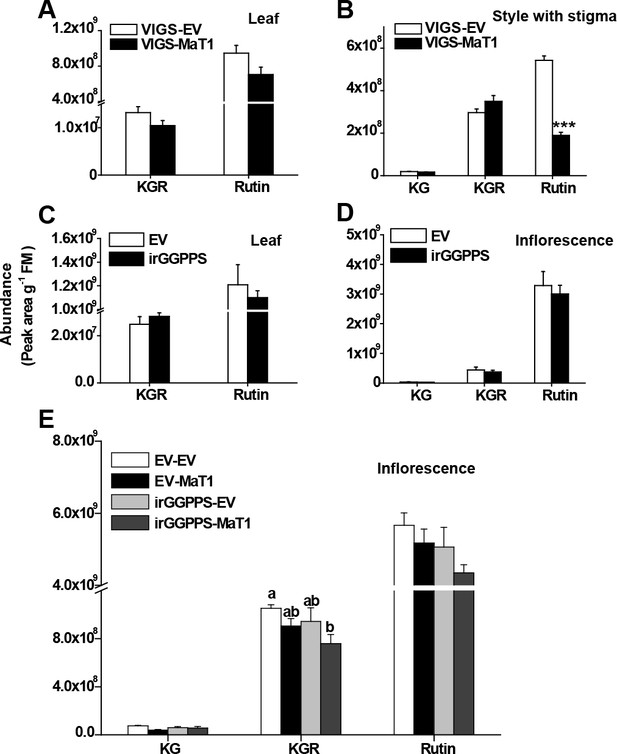
Silencing NaMaT1 and NaGGPPS does not affect flavonoids in different tissues, except that VIGS of NaMaT1 reduces rutin in the style.
Different tissue samples were harvested and extracted as described for DTG profile analysis. Relative abundance (mean + SE; n = 9 – 10) of different flavonoids are shown for leaf (A) and style with stigma (B) of VIGS-MaT1 and VIGS-EV plants. Relative abundance (mean + SE; n = 5 – 7) of different flavonoids are shown for leaf (C) and inflorescences (D) of irGGPPS plants and EV control. (E) Relative abundance (mean + SE; n = 9 – 10) of different flavonoids in inflorescences of VIGS plants in the background of EV and irGGPPS. KG, kaempferol-3-O-glucoside; KGR, kaempferol-3-O- rhamnosyl glucoside; Rutin, quercetin-3-O- rhamnosyl glucoside (QGR). Asterisks indicate significant differences between EV and VIGS-MaT1 plants (***p<0.001; Student’s t-test). Different letters indicate significant differences between different genotypes (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
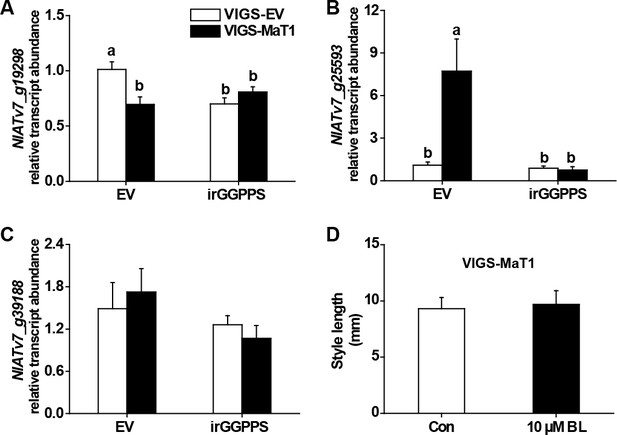
Brassinosteroid does not contribute to the VIGS-MaT1 short style.
(A–C) Relative transcript abundance (mean + SE; n = 5) of PveCYP734A50 homologs in N. attenuata were analyzed by qRT-PCR in styles dissected from flowers one day before anthesis. (D) Style lengths (mean + SE; n = 8) of freshly opened VIGS-MaT1 flowers which had been injected with 0.1% DMSO aqueous solutions with 10 µM brassinolide (10 µM BL) or only 0.1% DMSO aqueous solution as controls (Con) two days before. Different letters indicate significant differences among different plant lines or treatments (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
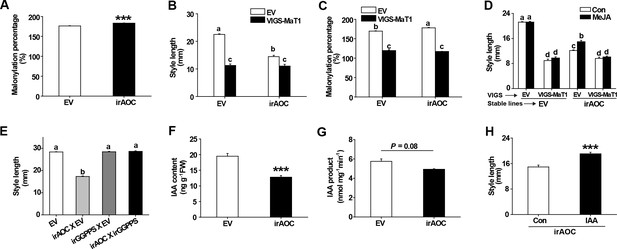
The short styles of jasmonate-deficient irAOC plants result from disturbed DTG malonylation status.
(A) DTG malonylation percentage (mean + SE; n = 5) was calculated based on DTGs of styles dissected from flowers one day before anthesis. (B) Style lengths (mean + SE; n = 10) were measured from freshly opened flowers of VIGS-EV and VIGS-MaT1 silencing in the background of two stably transformed lines, EV or irAOC, as indicated. (C) Malonylation percentages (mean + SE, n = 3) were calculated based on DTGs in styles dissected from flowers one day before anthesis of the four genotypes as described in (B). (D) Mean style lengths (mean + SE; n = 10) were measured from freshly opened flowers of the four genotypes, which had been treated with lanolin paste with MeJA (MeJA) or only lanolin paste as control (Con), 2 days previously. (E) Style lengths (mean + SE; n = 5) were measured from freshly opened flowers of the four genotypes, which were from crosses of EV or irAOC with EV or irGGPPS. (F) IAA levels (mean + SE; n = 5) were analyzed from the same samples as in (A). (G) IAA (mean + SE; n = 3) was quantified from in vitro enzyme activity assay products, in which the enzyme activity is from crude protein extracted from styles as in (A). (H) Style lengths (mean + SE; n = 10 – 16) of freshly opened irAOC flowers which had been injected with 0.1% DMSO aqueous solutions with 10 µM IAA (IAA) or only 0.1% DMSO aqueous solution as controls (Con) 2 days before. Asterisks indicate significant differences between different genotypes or treatments (***p<0.001; Student’s t-test). Different letters indicate significant differences among different plant lines or treatments (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
-
Figure 6—source data 1
The short styles of jasmonate-deficient irAOC plants result from disturbed DTG malonylation status.
- https://doi.org/10.7554/eLife.38611.033
-
Figure 6—source data 2
DTG malonylation status and style length are affected by silencing a JA biosynthesis gene (AOC), and are partly recovered by exogenous applications of MeJA.
- https://doi.org/10.7554/eLife.38611.034
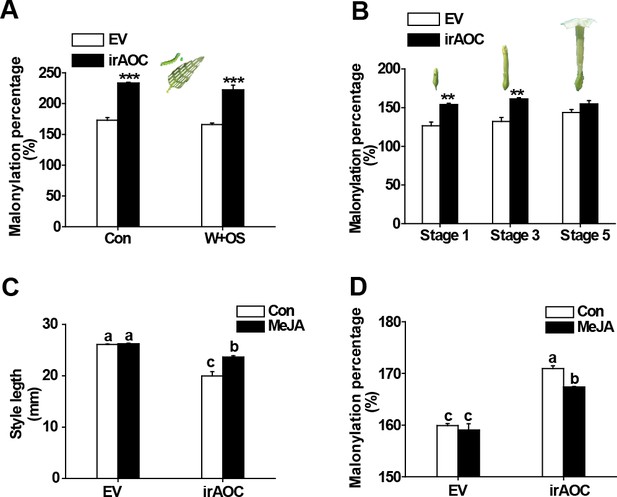
DTG malonylation status and style length are affected by silencing a JA biosynthesis gene (AOC), and are partly recovered by exogenous applications of MeJA.
(A) Mean malonylation percentage (mean + SE; n = 5) of DTGs in EV and irAOC leaves, which were either untreated (Con) or treated with wounding and M. sexta oral secretions (W + OS) and harvested after 3 days. (B) Mean malonylation percentage (mean + SE; n = 5) of DTGs in different stage flower buds of EV and irAOC plants. (C) Mean style lengths (mean + SE; n = 5) were measured at full anthesis from EV and irAOC flowers, which had been treated with lanolin (Con) or lanolin and MeJA (MeJA) 2 days prior to anthesis. (D) Mean malonylation percentage (mean + SE; n = 5) of DTGs in EV and irAOC flower buds which were treated with lanolin (Con) or MeJA 2 days before. Asterisks indicate significant differences between EV and irAOC (**p<0.01; ***p<0.001; Student’s t-test). Different letters indicate significant differences among different lines or treatments (p<0.05, one-way ANOVA followed by Tukey’s HSD post-hoc tests).
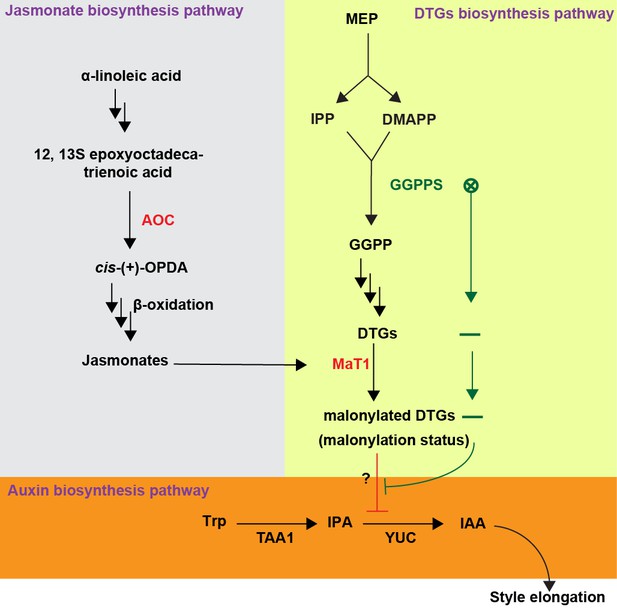
Model summarizing how DTGs malonylation affects style elongation of N. attenuata.
Geranylgeranyl diphosphate synthase (GGPPS) forms GGPP from the MEP pathway products IPP and DMAPP. This GGPP is used for DTG biosynthesis and a certain portion of these DTGs are decorated with malonic acid moieties by NaMAT1. When AOC or MaT1 are silenced (red), this disturbs the uniformity of DTG malonylation, resulting in compounds which inhibit the rate-limiting step of IAA biosynthesis, YUC catalytic activity. When GGPPS is silenced (green), leading to DTG deficiency, the suppression of YUC catalytic activity is alleviated. MEP, Methylerythritol 4-phosphate pathway; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Nicotiana attenuata) | NaMaT1 | PRJNA355166 | XM_019403695.1 | |
| Gene (N. attenuata) | NaMaT2 | PRJNA355166 | XR_002066055.1 | |
| Gene (N. attenuata) | NaMaT3 | PRJNA355166 | XM_019382488.1 | |
| Gene (N. attenuata) | NIATv7_g21823 | Nicotiana attenuata Data Hub | NIATv7_g21823 | |
| Gene (N. attenuata) | NIATv7_g39356 | Nicotiana attenuata Data Hub | NIATv7_g39356 | |
| Strain, strain background (Agrobacterium tumefaciens) | GV1301 | DOI: 10.1007/ 978-1-62703-278-0_9 | GV1301 | |
| Strain, strain background (Escherichia coli) | BL21 (DE3) | New England Biolabs inc. | catalog#: C2527I | |
| Genetic reagent (N. attenuata) | Empty vector (EV) | DOI: 10.1371/ journal.pone. 0001543 | EV | |
| Genetic reagent (N. attenuata) | irGGPPS | DOI: 10.1105/ tpc.109.071449 | irGGPPS | |
| Genetic reagent (N. attenuata) | irAOC | DOI: 10.1073/ pnas.1200363109 | irAOC | |
| Transfected construct (tobacco rattle virus) | pBINTRA6/pTV00 | DOI: 10.1007/ 978-1-62703-278-0_9 | pBINTRA6/pTV00 | |
| Recombinant DNA reagent | Gateway vector pDEST15 | Invitrogen | catalog#: 11802014 | |
| Sequence- based reagent | Oligonucleotides | Sigma-aldrich | Supplied in Supplementary file 3 | |
| Peptide, recombinant protein | GST-NaMaT1 | This paper | ||
| Peptide, recombinant protein | GST-NaMaT2 | This paper | ||
| Peptide, recombinant protein | GST-NaMaT3 | This paper | ||
| Peptide, recombinant protein | GST-NIATv7_g21823 | This paper | ||
| Peptide, recombinant protein | GST-NIATv7_g39356 | This paper | ||
| Commercial assay or kit | SuperScript First-Strand Synthesis System for RT-PCR | Invitrogen | catalog#: 11904018 | |
| Chemical compound, drug | TRIzol TM Reagent | Invitrogen | catalog#: 15596026 | |
| Chemical compound, drug | Indole-3- pyruvic acid | Sigma-aldrich | CAS:392-12-1 | |
| Chemical compound, drug | Glutathione- Sepharose 4B | GE Healthcare | GE17-0756-01 | |
| Software, algorithm | MEGA6 | MEGA | http://www.megasoftware.net/ | |
| Software, algorithm | SPSS statistic 17.0 | SPPS inc. | http://www-01.ibm.com/software/analytics/spss/ |
Additional files
-
Supplementary file 1
Gene accession numbers used in this study.
- https://doi.org/10.7554/eLife.38611.036
-
Supplementary file 2
The transcripts per million (TPM) values of DTGs biosynthesis pathway genes, NaStyle2.1 and NaYUC-like, from RNAseq analysis.
- https://doi.org/10.7554/eLife.38611.037
-
Supplementary file 3
DNA primers used in this study.
- https://doi.org/10.7554/eLife.38611.038
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38611.039





