Distinct and evolutionary conserved structural features of the human nuclear exosome complex
Figures
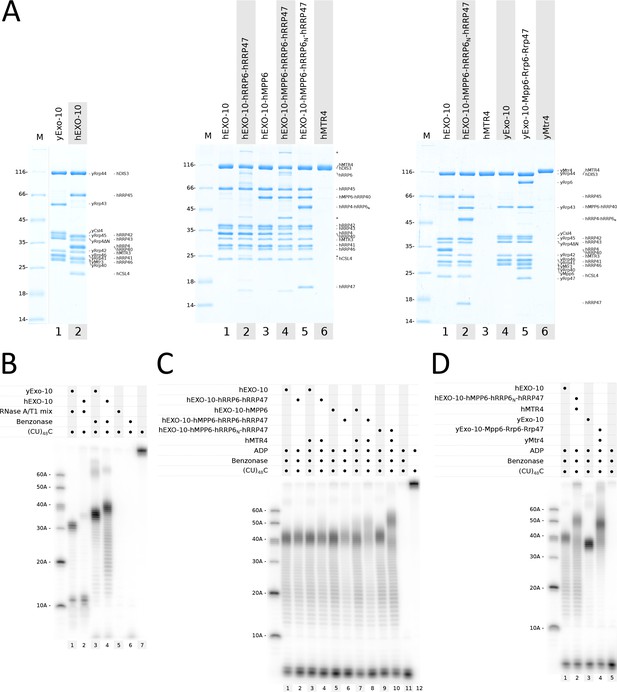
Biochemical analysis of the RNA-binding paths in yeast and human nuclear exosome complexes.
(A) Coomassie-stained 12.5% SDS-PAGE gels showing reconstituted yeast and human exosome complexes and subunits used in the assays in panels (B–D). All samples correspond to the pooled peak fractions from size exclusion chromatography. hMPP6-hRRP40 and hRRP4-hRRP6N indicate the genetically linked fusion proteins, as described in the text. Undefined contaminants and degradation products from the hRRP6-hRRP47 preparation are indicated with asterisks.(B–D) RNase protection assays showing the RNA fragments obtained upon RNase treatment with 32P body-labeled (CU)48C 97-mer RNA in the presence of the indicated protein complexes. After incubation with RNase A/T1 or with benzonase (Serratia marcescens endonuclease), the reactions products were analyzed by electrophoresis on a 12% acrylamide and 7M urea gel, followed by phosphorimaging. Protein concentrations were 1 μM (panel B) or 500 nM (panels C, D). Substrate concentration was 250 nM. The outer left lanes were loaded with size markers. Note that the size of fragments obtained in these in vitro assays is not an absolute measure of the length of the RNA-binding channel but is relative to the RNase used (panel B, compare lanes 1 and 3).
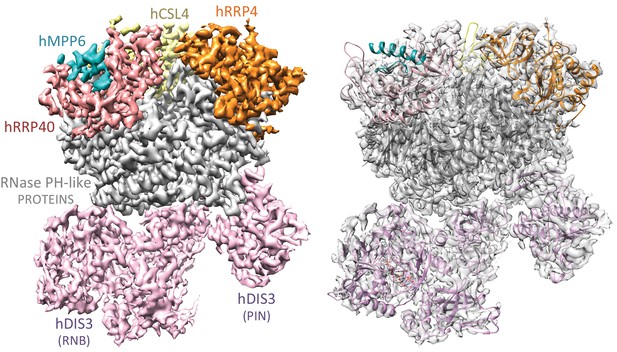
Cryo-EM structural analysis of a human nuclear exosome core.
Cryo-EM surface representation (left panel) and cryo-EM density (right panel) of hEXO-10cat-hMPP6 with the corresponding atomic coordinates of the individual exosome subunits. The hEXO-9 barrel comprises a base ring of 6 RNase PH-like subunits (all shown in gray) and a cap ring of 3 S1/KH-like proteins (shown at the top in yellow, orange and salmon). The PIN and exoribonuclease regions of hDIS3 are indicated (shown in light pink). The hMPP6 cofactor (shown in teal) is bound to the protein hRRP40.
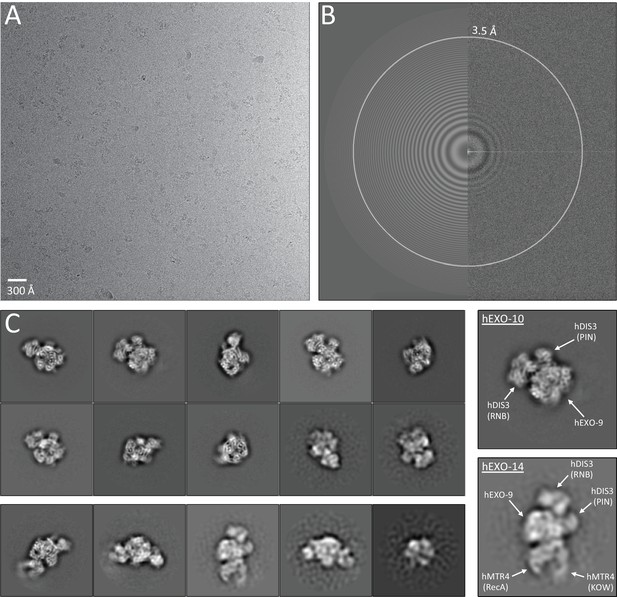
Cryo-EM data collection and 2D classification.
(A) An example of the cryo-EM micrograph collected on a FEI Titan Krios operated at 300 kV, equipped with a K2 Summit camera. The scale bar corresponds to 300 Å. (B) Thon-ring image of the same micrograph as in (A) following the contrast transfer function estimation with GCTF. Zeros between the Thon rings in the experimental image (right half) coincide with the ones in the model (left half). (C) Left panels represent the reference-free 2D class averages from the hEXO-10 (top) and the hEXO-14 (bottom). Two panels on the right highlight domain organization of both hEXO-10 (top) and hEXO-14 (bottom).
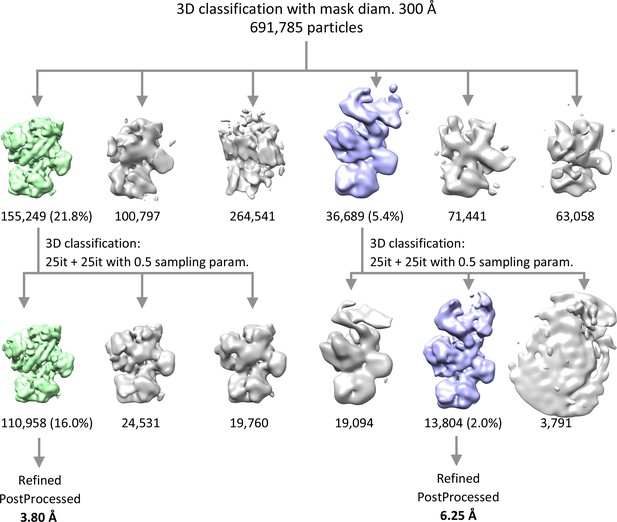
Cryo-EM data processing scheme.
The 691,785 particles from the 2D classification were initially 3D classified into six classes, using 300 Å mask. Two best classes representing the hEXO-10 and the hEXO-14 complexes were subsequently aligned and 3D classified further separately into three classes each, using finer sampling parameters. The best 3D classes of the hEXO-10 and the hEXO-14 were autorefined and postprocessed.
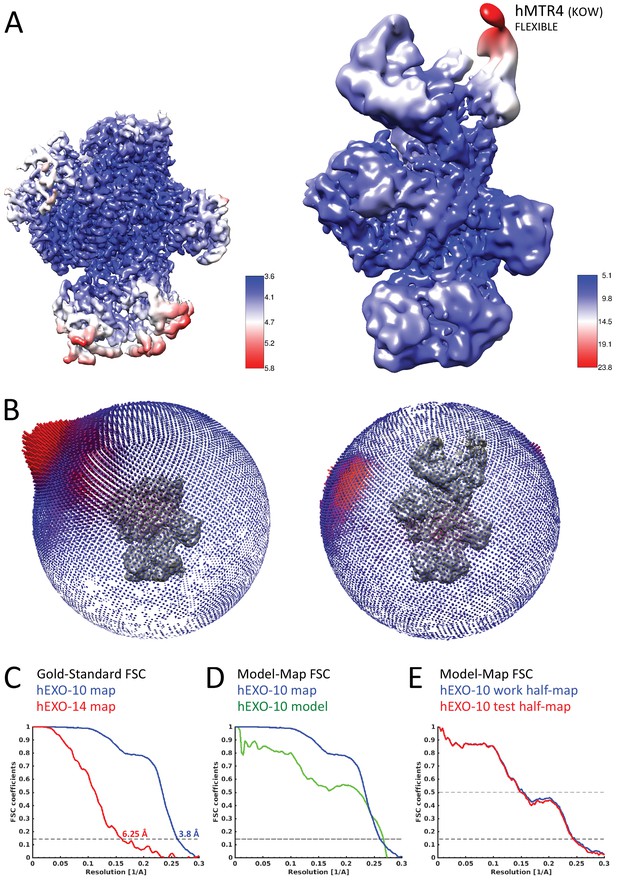
Cryo-EM data quality.
(A) Local resolution analysis of both hEXO-10 (left) and hEXO-14 (right). Maps show variation in local resolution, as estimated by RELION. The hEXO-10 map shows high resolution in most of the areas, with the hEXO-9 barrel resolved up to 3.6 Å. Solvent exposed regions of hDIS3 and the hRRP40-hMMP6 interface have lower resolution. The same is true for the hEXO-14 map. The hMTR4 helicase has lower resolution of around 10 Å. The hMTR4 KOW domain is the worst resolved part, most likely due to its flexibility. (B) Angular distribution of the hEXO-10 (left) and the hEXO-14 (right) particles used for the autorefinement. (C) Fourier shell correlation (FSC) curves for the hEXO-10 (blue) and hEXO-14 (red). The gold standard FSC value of 0.143 and resolutions are indicated. (D) FSC from the RELION autorefine procedure (blue) and Model vs. map FSC for the final Phenix real space refined model of the hEXO-10 reconstruction (green). (E) The model was probed for over-fitting by randomly perturbing the atoms by 0.5 Å and refinement against the first of the two independent half-maps (work half-map, blue). The resulting refined model was then used to calculate Model-Map FSCs against the second half-map (test half-map, red) that was not used for refinement. The FSC_work and FSC_test curves show an excellent agreement in the entire resolution range, validating the entire structure for over-fitting.
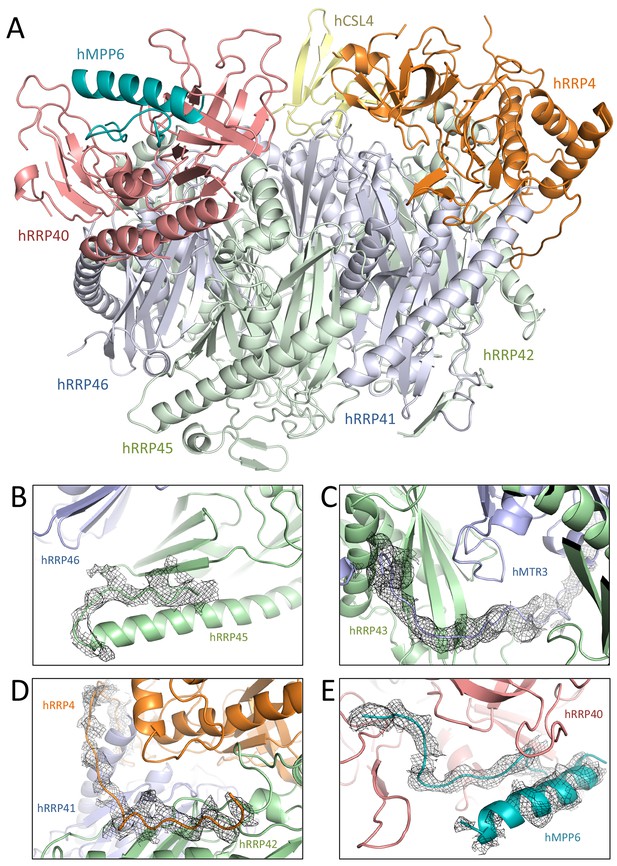
Human and yeast exosome cores: extensive similarities of the central scaffolds.
(A) Structure of the hEXO-9 barrel from the cryo-EM reconstruction bound to hMPP6 (teal). In hEXO-9, the cap proteins are shown in the same colors as in panel A and the base proteins are in light blue (for the Rrp41-like proteins hRRP41, hRRP46 and hMTR3) and light green (for the Rrp42-like proteins hRRP42, hRRP43 and hRRP45). The definition of Rrp41-like and Rrp42-like follows the original description in (Lorentzen et al., 2005). The superposition of the human and yeast scaffolds is shown in Figure 3—figure supplement 1. (B–D) Zoom-ins at specific regions of human hEXO-9 described in text, with cryo-EM density superposed. (E) Zoom-in at hMPP6 fragment bound to hRRP40, with cryo-EM density superposed.
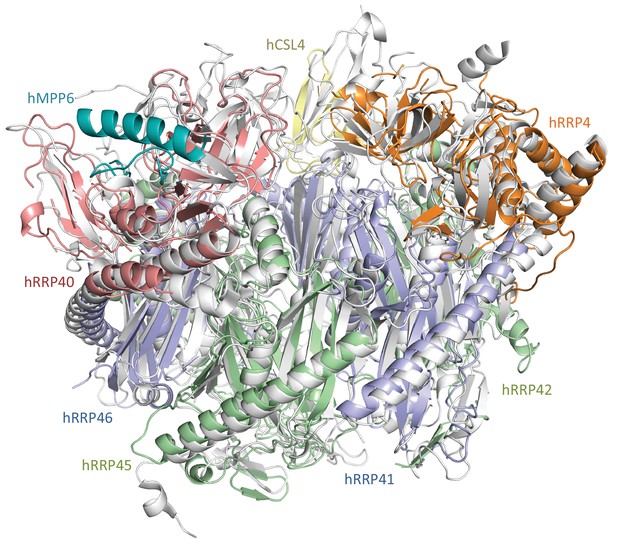
Similar scaffolds of yeast and human exosome cores.
The cryo-EM structure of hEXO-9 (in the same colors as in Figure 3) is shown superposed to the structure of yExo-9 (Makino et al., 2015); PDB: 5c0w.
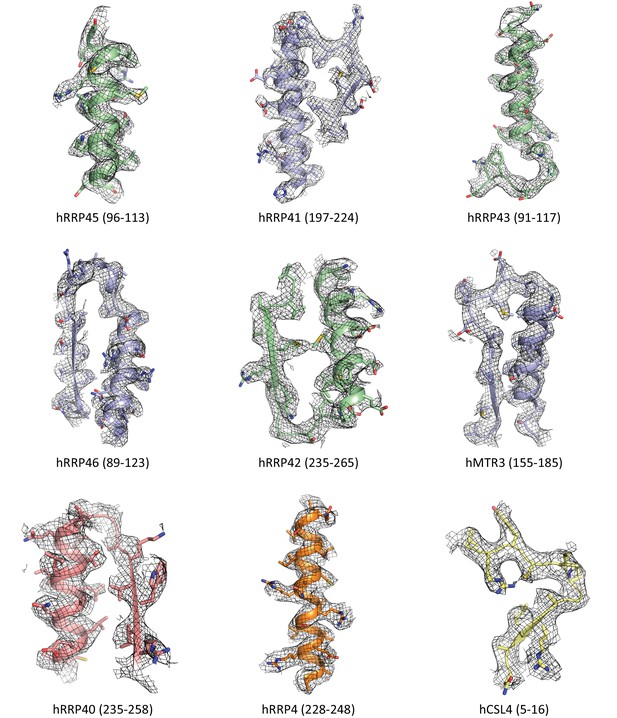
Zoomed in views of the hEXO-9 subunits.
Representative parts of the hEXO-9 core exosome complex from the hEXO-10 EM density map, showing secondary structure elements and resolved side chains. Subunit names and highlighted sequences are marked below corresponding views.
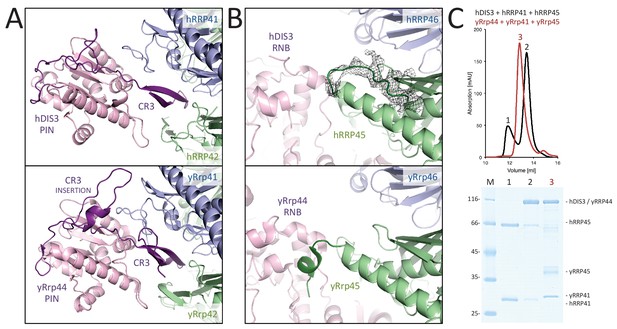
Human and yeast exosome cores: differential binding to the exoribonuclease.
(A-B) Interactions made by the PIN region (A) and the RNB region (B) of the exoribonuclease (pink) with the proteins of the central scaffold (shown in the same colors as in Figure 3). The figures show the comparisons between the human cryo-EM structure (upper panels) and previous yeast X-ray structures (lower panels) (Makino et al., 2015, 2013). The human complex shows fewer interactions at the structural level as compared to the yeast complex. (C) Size exclusion chromatography experiments showing the weaker binding of hDIS3 to hRRP41-hRRP45 as compared to that of the yeast orthologues.
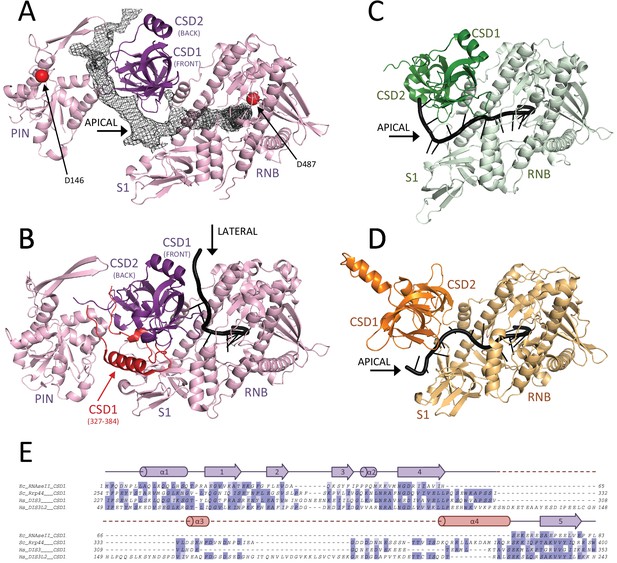
RNA binding to hDIS3 is more similar to hDIS3L2 and RNase II than to yRrp44.
(A) The structure of hDIS3 from the cryo-EM study is shown with the density corresponding to the bound RNA, segmented from the autorefined hEXO-10 map. (B) The structure of the S. cerevisiae orthologue yRrp44 in the open conformation with an RNA molecule accessing the RNB active site through the lateral entry. CSD1 region which impairs apical entry through the CSD1-S1 route is depicted in red (Makino et al., 2015). (C) The crystal structures of the paralogue mouse DIS3L2 (Faehnle et al., 2014) and (D) of the E. coli RNase II (Frazão et al., 2006). All structures are shown in the same orientation after optimal superposition of their RNB domains. In all panels the RNB and S1 domains form a rather rigid module and are shown in lighter colors, while the CSD1 and CSD2 domains are shown in darker colors. (E) Sequence alignment of the CSD1 domains from E. coli RNase II, S. cerevisiae yRrp44, human DIS3, and human DIS3L2. The secondary structure of the yRrp44 CSD1 region that impairs apical entry through the CSD1-S1 route is depicted in red.
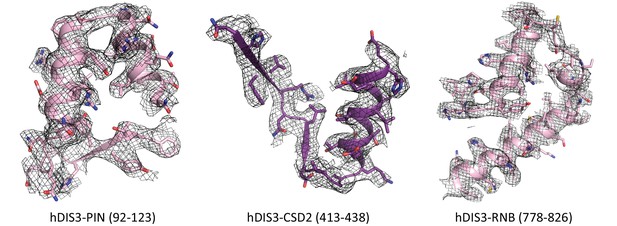
Zoomed in views of the hDIS3.
Representative parts of the hDIS3 PIN, CSD2, and RNB domains from the hEXO-10 EM density map, showing secondary structure elements and resolved side chains. Domains and highlighted sequences are marked below corresponding views.
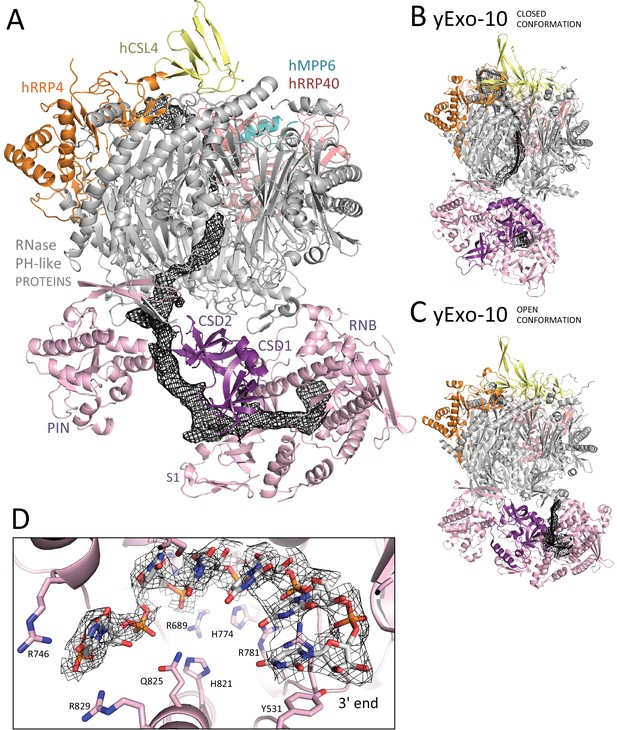
The RNA channel paths in the human and yeast exosome core complexes.
(A) Cartoon representation of the hEXO-10cat-hMPP6 cryo-EM structure with highlighted the density corresponding to a single-stranded RNA traversing the exosome channel, segmented from the autorefined hEXO-10 map. (B-C) Comparison with the two yeast exosome crystal structures with RNA bound in the channel path (B) (Makino et al., 2015), PDB: 5c0x) and in the direct path (C) (Makino et al., 2015), PDB: 5c0w). The structures are shown in the same orientation after superposition of the yExo-9 barrels. The open and closed conformations of yRrp44 are indicated. (D) Zoom-in of at the active site of hDIS3. Meshed density from the hEXO-10 map surrounds six nucleotides built in the atomic model. Conserved residues are highlighted, and correspond to similar interactions in yRrp44 (Lorentzen et al., 2008; Makino et al., 2013).

Structural conservation of human and yeast nuclear exosome cofactors.
(A) On the left and central panels is the surface representation of the hEXO-14cat cryo-EM structure. On the right is a cartoon representation of the yExo-14 structure from (Schuller et al., 2018, PDB: 6fsz), shown in the same orientation and color coding. (B) On the left is a growth assay of wild-type and mutant mtr4 strains. Endogenous MTR4 was replaced with wild-type or mutant mtr4-EGFP fusions. Cells were grown to early exponential phase, and serial dilutions were spotted onto 5-fluoroorotic acid (FOA) medium or control plate. Medium containing FOA selects for the loss of the rescue vector. SC, synthetic complete medium; YPD, yeast extract peptone adenine dextrose; FOA, 5-fluoroorotic acid. ΔSK corresponds to an arch-less mutant of yMtr4 (Falk et al., 2014) On the right is the analysis of the expression levels of wild type yMtr4-EGFP and mutants by Western blotting using an anti-GFP antibody.
-
Figure 7—source data 1
Detailed information of the yeast strains used in this study.
- https://doi.org/10.7554/eLife.38686.016
Tables
Statistics of the hEXO-10cat-hMPP6 cryo-EM atomic model.
https://doi.org/10.7554/eLife.38686.010| hEXO-10cat-hMPP6 | |
|---|---|
| Data collection | |
| Microscope | Titan Krios |
| Camera | Gatan K2 Summit |
| Magnification | 105,000 x |
| Voltage (kV) | 300 |
| Electron dose (e-/Å2) | 46.9 |
| Dose rate (e-/pixel/s) | 8.55 |
| Defocus range (μm) | 0.5–3.5 |
| Pixel size | 1.35 |
| Reconstruction | |
| Micrographs collected | 8047 |
| Particles in 3D classification | 691,785 |
| Particles in final refinement | 110,958 |
| Refinement | |
| Resolution (Å) | 3.8 |
| No. atoms | 22117 |
| Protein | 21997 |
| RNA | 120 |
| Map sharpening B-factor | −153 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 0.926 |
| Ramachandran plot | |
| Favored (%) | 94.2 |
| Allowed (%) | 5.7 |
| Rotamers ouliers (%) | 0.65 |
| MapCC Global | 0.79 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38686.017





