Generation of a versatile BiFC ORFeome library for analyzing protein–protein interactions in live Drosophila
Figures
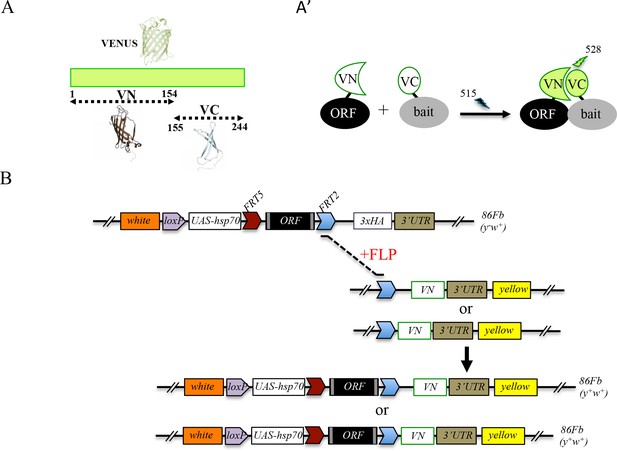
Generation of a Gal4 inducible library compatible with Venus-based BiFC in Drosophila.
(A-A’) Principle of the Venus-based BiFC between a candidate ORF (Open Reading Frame) and a bait protein fused to the N- (VN) or C-terminal (VC) fragment of Venus, respectively. Excitation and emission wavelengths are indicated. (B) Principles of Flippase (FLP)/FRT-mediated recombination to swap the C-terminal 3xHA tag of the ORF with the original VN or new VN-short tag line. Genetic crosses and selection procedure are described in (Bischof et al., 2013). Note that the UAS-ORF-HA and resulting UAS-ORF-VN are located on the third chromosome (86Fb). See also Figure 1—figure supplement 1 and Supplementary file 1.
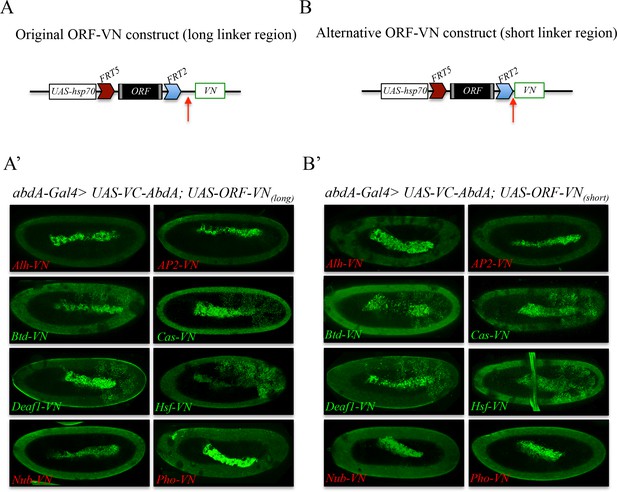
Comparison of Venus-based BiFC when using ORFs swapped with the original VN or VN-short tag line.
(A) Schematic representation of an ORF fused to VN with the original linker region (red arrow). (A’) Illustrative confocal captures of BiFC resulting from the co-expression of ORFs fused to VN with the original linker region and the Hox protein AbdominalA (AbdA) fused to the complementary VC fragment. (B) Schematic representation of an ORF fused to VN with the new short linker region (red arrow). (B’) Illustrative confocal captures of BiFC resulting from the co-expression of ORFs fused to VN with the new short linker region and the Hox protein AbdominalA (AbdA) fused to the complementary VC fragment. Fusion proteins are expressed with the abdA-Gal4 driver and BiFC is shown in the epidermis of stage 10 embryos. Note that the amnioserosa inside the embryo displays prominent autofluorescence. ORFs that are negative with AbdA are highlighted in red.
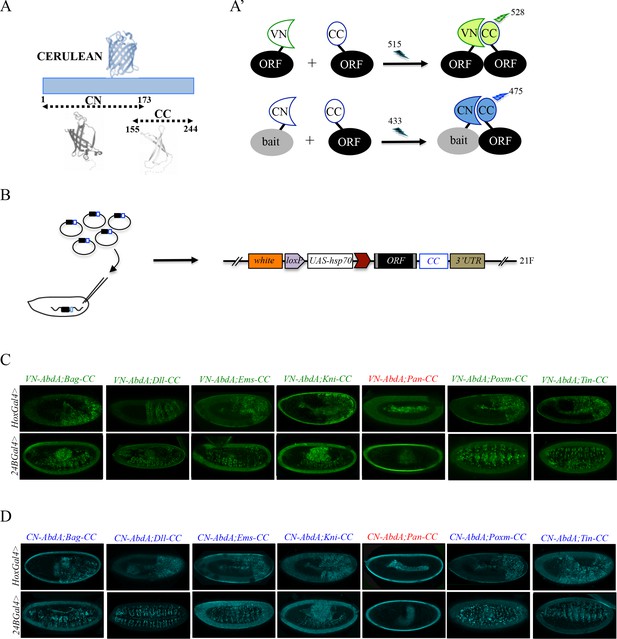
Generation of a Gal4 inducible library compatible with Venus- and Cerulean-based BiFC in Drosophila.
(A-A’) Principle of bicolor BiFC by using the complementation property between the C-terminal fragment of the blue fluorescent protein Cerulean (CC) and the N-terminal fragment of Venus (VN) or Cerulean (CN). Excitation and emission wavelengths are indicated. (B) Principle of the generation of the UAS-ORF-CC library at the 21F genomic locus. See also Materials and methods. (C-D) Illustrative confocal captures of Venus-based BiFC obtained from different ORF-CCs and VN-AbdA (C) or CN-AbdA (D) interaction partners, as indicated. Fusion proteins are expressed with the abdA-Gal4 (upper panels) or 24B-Gal4 (lower panels) driver and BiFC is observed in the epidermis (stage 10/11) or somatic mesoderm (stage 14), respectively. Note that the amnioserosa, the gut inside the embryo and the vitelline membrane around the embryo display strong autofluorescence. Absence of interaction with Pangolin (Pan) is highlighted in red. See also Figure 2—figure supplements 1 and 2 and Supplementary file 2 and 3.
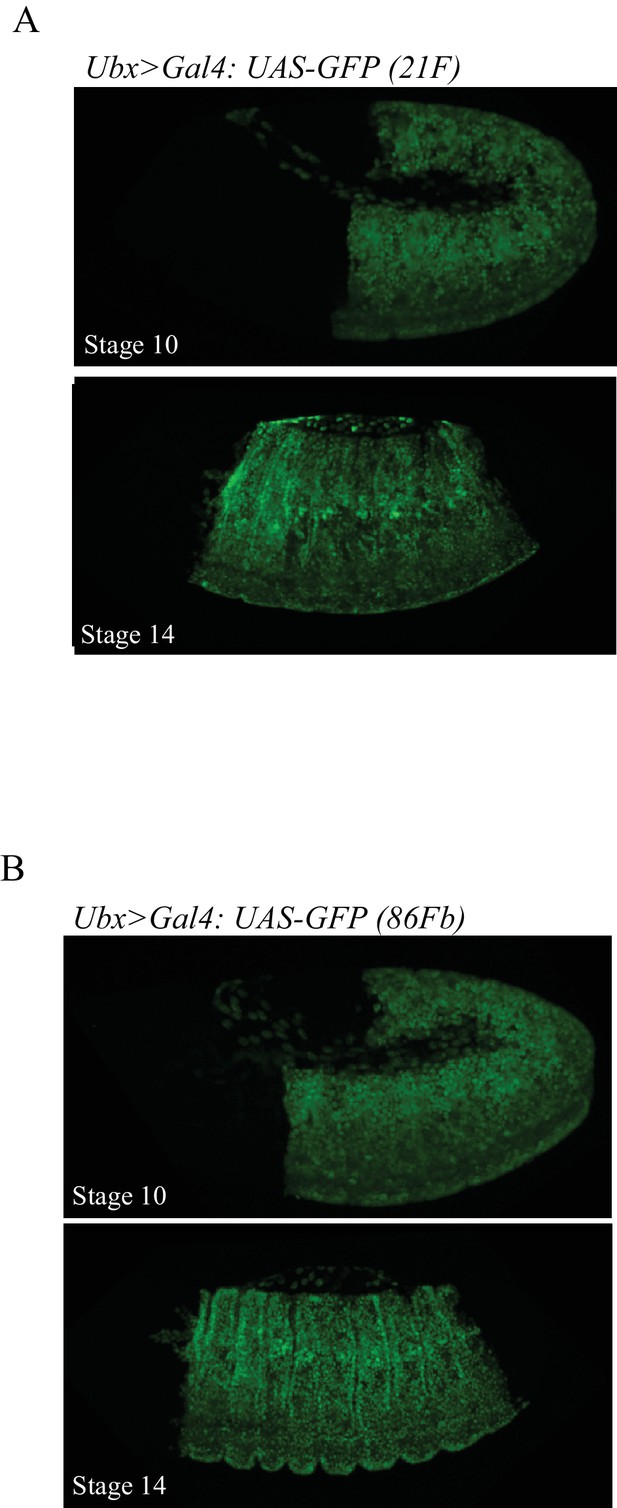
The 21F and 86Fb attP sites lead to similar expression levels.
. Illustrative confocal capture of UAS-driven GFP inserted in the 21F (A) or 86Fb (B) attP sites with the Ubx-Gal4 driver. Two different stages are shown.
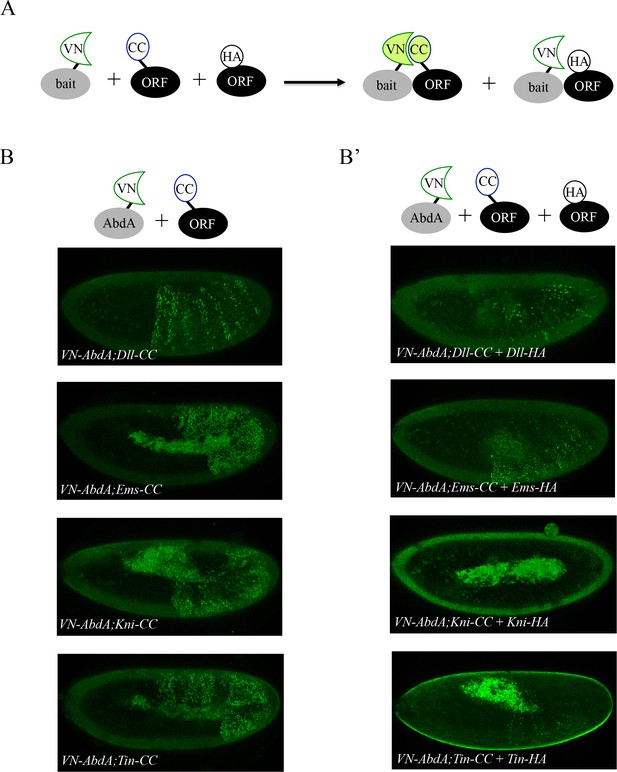
Co-expression of non-complementing ORF-3xHA can compete against BiFC signals obtained from ORF-CC and VN-AbdA constructs.
(A) Principle of the BiFC competition experiment with co-expression of a non-complementing HA-tagged ORF. (B) Illustrative confocal captures of BiFC between VN-AbdA and the different ORF-CC constructs, as indicated. (B’) Illustrative confocal captures of BiFC between VN-AbdA and the same ORF-CC constructs in the presence of the corresponding ORF-HA. Constructs are expressed with the abdA-Gal4 driver and BiFC is analysed in the epidermis of stage 10/11 embryos. The co-expression of non-complementing ORF-HA strongly affects BiFC in all cases.
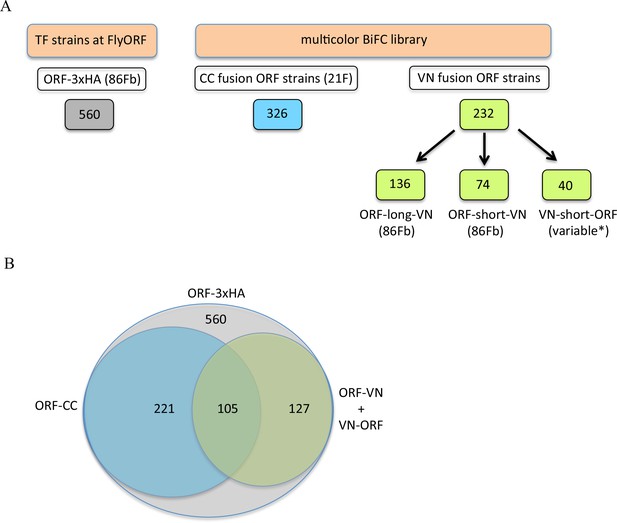
The TF-3xHA and multicolour BiFC libraries.
(A) Number of transcription factors (TFs) tagged with three repeats of the hemagglutinin (3xHA) epitope at the 3’ end and multicolour BiFC fly strains at FlyORF. Various insertion sites were used for the VN-short-ORF constructs (Baëza et al., 2015). Some of the 232 TF strains exist in more than one VN version (see also Supplementary file 1). (B) Distribution of the multicolour BiFC fly lines compared to the TF-3xHA library.
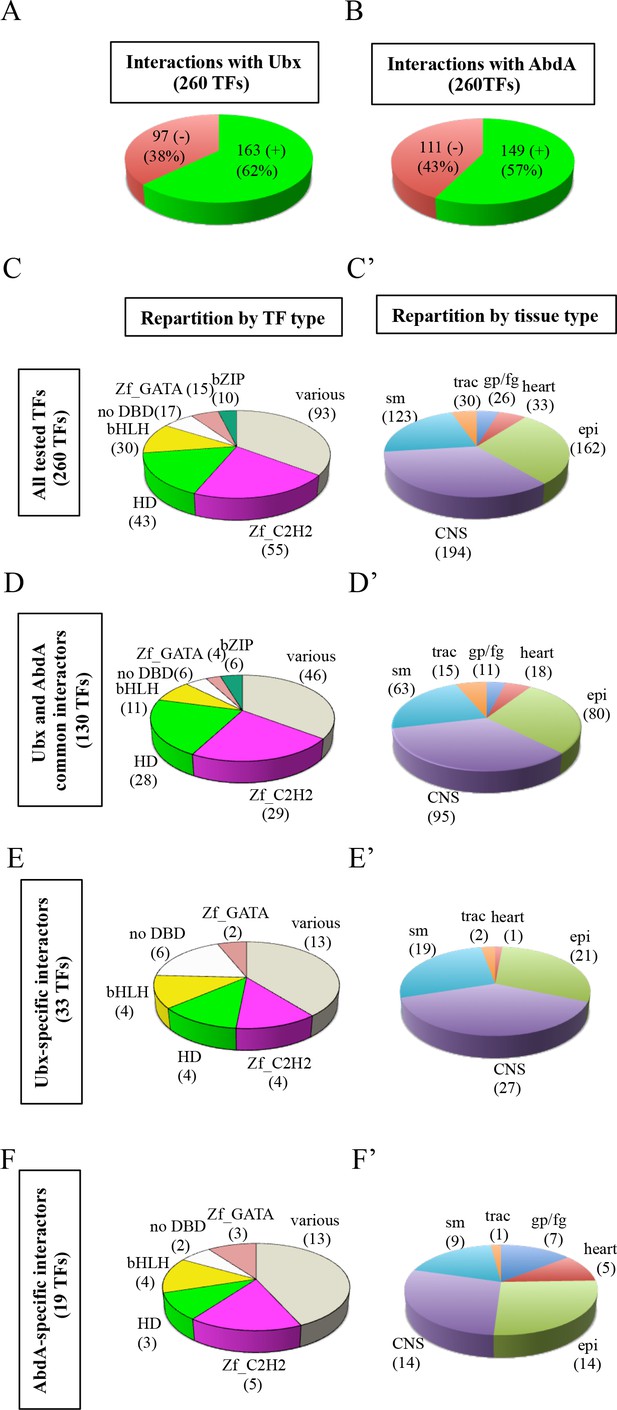
Using the multicolor BiFC library for a large-scale interaction screen with Ubx and AbdA in the live Drosophila embryo.
(A) Number of TFs that were positive (green) or negative (red) with Ubx. (B) Number of TFs that were positive (green) or negative (red) with AbdA. (C) Repartition of the different families among the 260 TFs tested with the Hox proteins. A specific color code is attributed to each TF family. Families with the highest number of tested TFs are represented (Zinc fingers C2H2, Zf_C2H2; homeodomain, HD; basic helix-loop-helix, bHLH; no DNA-binding domain, no DBD; zinc fingers GATA, Zf_GATA; basic leucine zipper, bZIP). 36 other different families containing one to eight TF representatives are present in the ‘various’ category. (D) Repartition of the TF families among the 130 positive interactions common to Ubx and AbdA. Note that the HD family is slightly enriched in this interactome. (E) Repartition of the TF families among the 33 Ubx-specific interactions. (F) Repartition of the TF families among the 19 AbdA-specific interactions. Note the absence of bZIP representatives in the Ubx- and AbdA-specific interactomes. (C’) Repartition of the expression profile of the 260 tested TFs in six different embryonic tissues: the somatic mesoderm (sm), trachea (trac), gonad primordium/fat body (gp/fb), heart, epidermis (epi) and central nervous system (CNS). Most of these TFs are expressed in several embryonic tissues. (D’) Tissue-type repartition of the expression profile of the 130 Ubx- and AbdA-positive interactors. (E’) Tissue-type repartition of the expression profile of the 33 Ubx-specific interactors. Note the absence of TFs expressed in the gp/fb. (F’) Tissue-type repartition of the expression profile of the 19 AbdA-specific interactors. Note the specific enrichment of TFs expressed in the gp/fb. See also Figure 4—figure supplements 1–10 and Supplementary file 4 and 5.
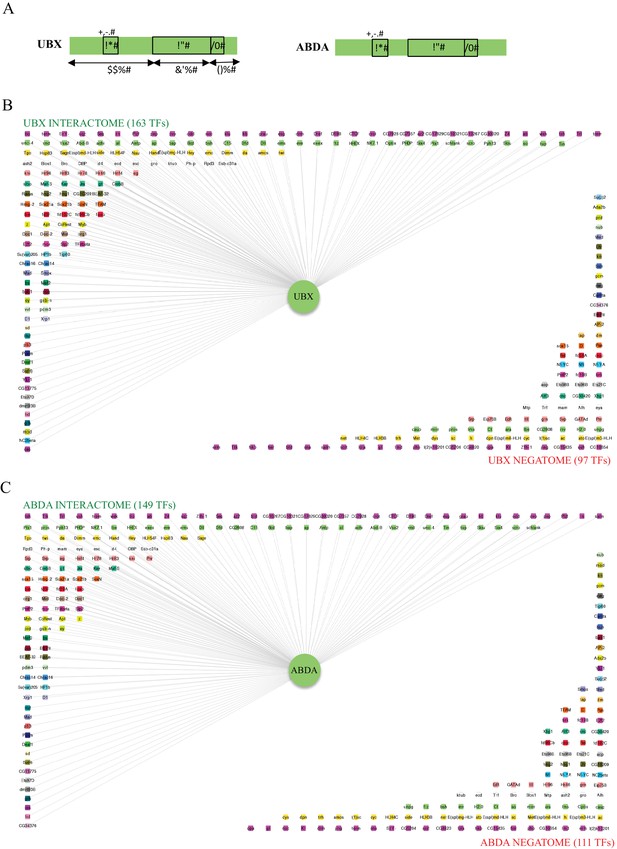
Interactomes and negatomes of Ubx and AbdA with the 260 tested TFs.
(A) Schematic representation of Ubx and AbdA. The homeodomain (HD), hexapeptide (HX) and UbdA (UA) motifs are indicated, as well as the percentage of identity of Ubx with AbdA along the corresponding regions. (B) Representation of the interactome (positive interactions) and negatome (negative interactions) of Ubx. (C) Representation of the interactome and negatome of AbdA. Compared to the negatome, HD-containing TFs (green boxes) are more enriched in each interactome. Color code is as in Figure 4 (for most represented TF families: Zinc fingers C2H2, Zf_C2H2: fuchsia; homeodomain, HD: light green; basic helix-loop-helix, bHLH: light orange; no DNA-binding domain, no DBD: white; zinc fingers GATA, Zf_GATA: pink; basic leucine zipper, bZIP: green). Networks are represented using Cytoscape 3.6 (Shannon et al., 2003).

Illustrative confocal captures of Venus-based BiFC between ORF-VN and VC-Ubx, as indicated.
Fusion proteins are expressed with the Ubx-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
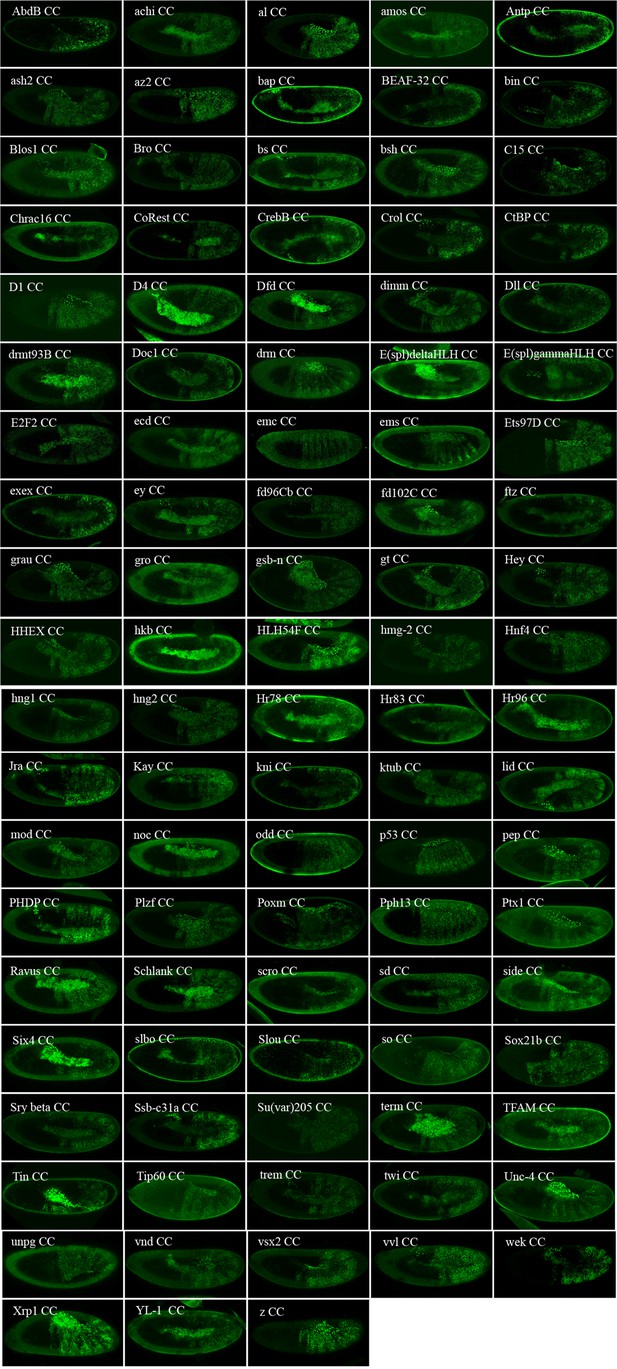
Illustrative confocal captures of Venus-based BiFC between ORF-CC and VN-Ubx, as indicated.
Fusion proteins are expressed with the Ubx-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
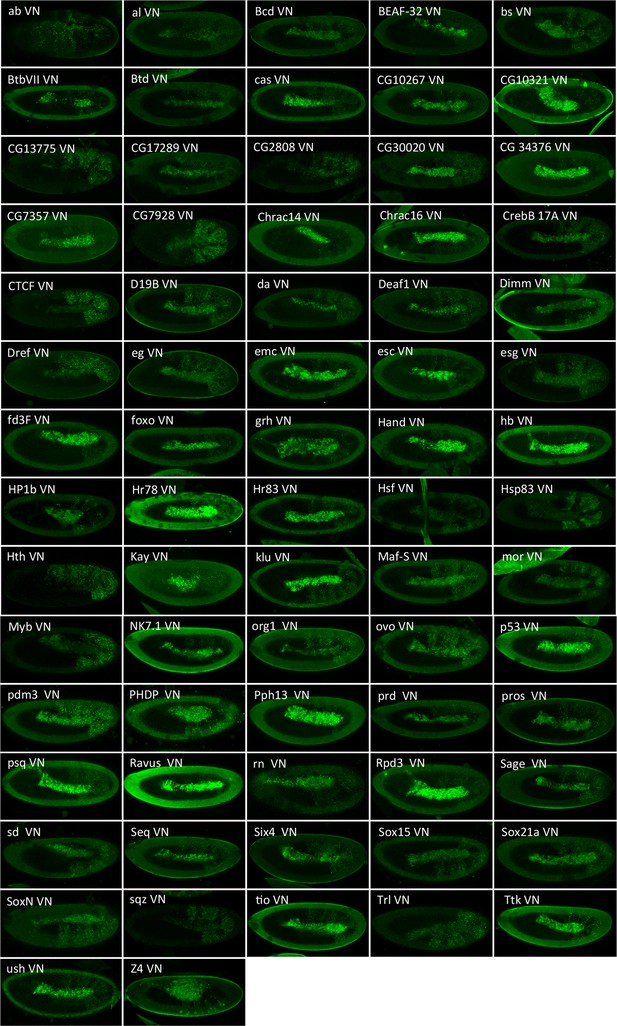
Illustrative confocal captures of Venus-based BiFC between ORF-VN and VC-AbdA, as indicated.
Fusion proteins are expressed with the abdA-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
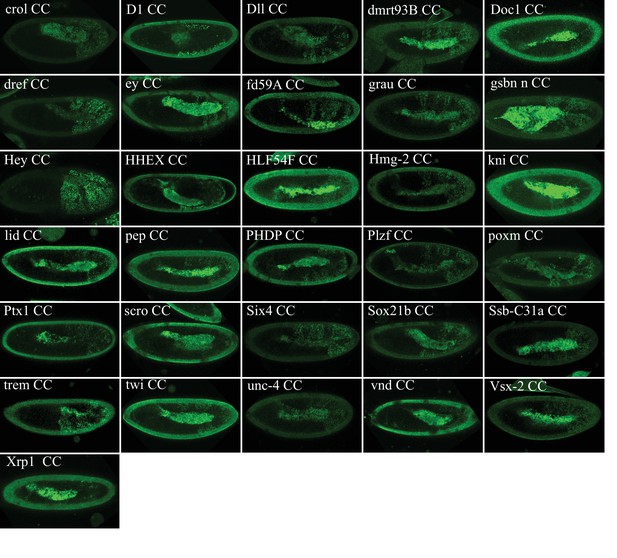
Illustrative confocal captures of Venus-based BiFC between ORF-CC and VN-AbdA, as indicated.
Fusion proteins are expressed with the abdA-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
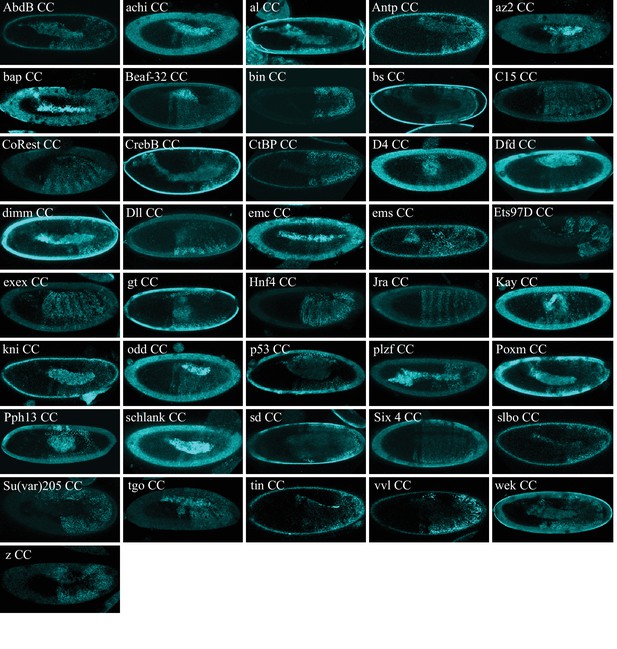
Illustrative confocal captures of Cerulean-based BiFC between ORF-CC and CN-AbdA, as indicated.
Fusion proteins are expressed with the abdA-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
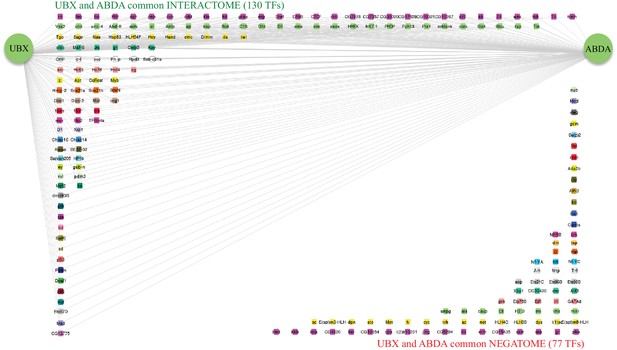
Representation of the common interactome and negatome between Ubx and AbdA.
Colour code and representation are as in Figure 4—figure supplement 1.
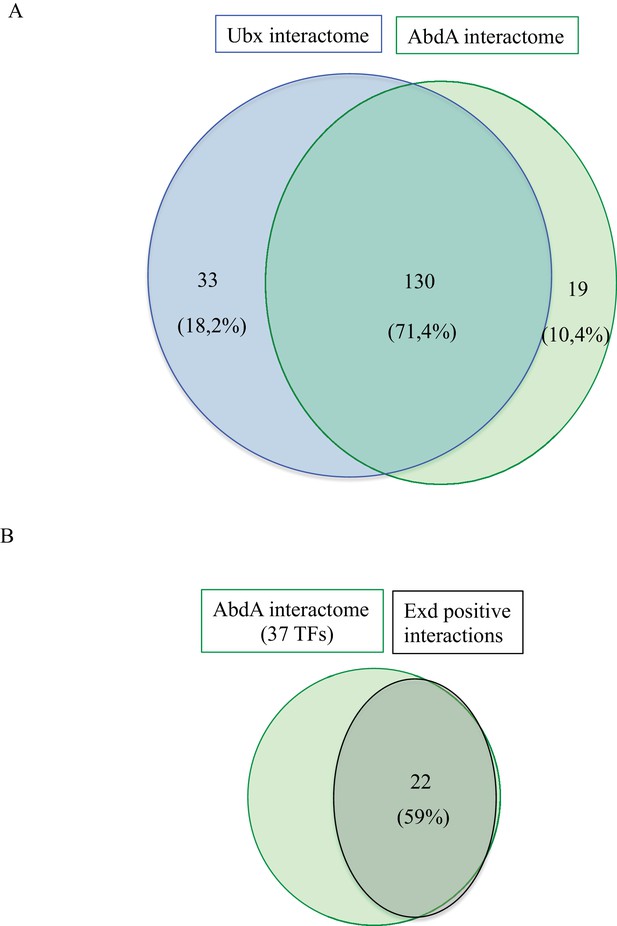
Comparison between Interactomes.
(A) Representation of the common and specific interactions among the interactomes of Ubx and AbdA. The corresponding percentage among the overall 184 positive interactions is indicated. (B) Representation of the proportion of positive interactions with Extradenticle (Exd) among 37 TFs that were positive with AbdA.
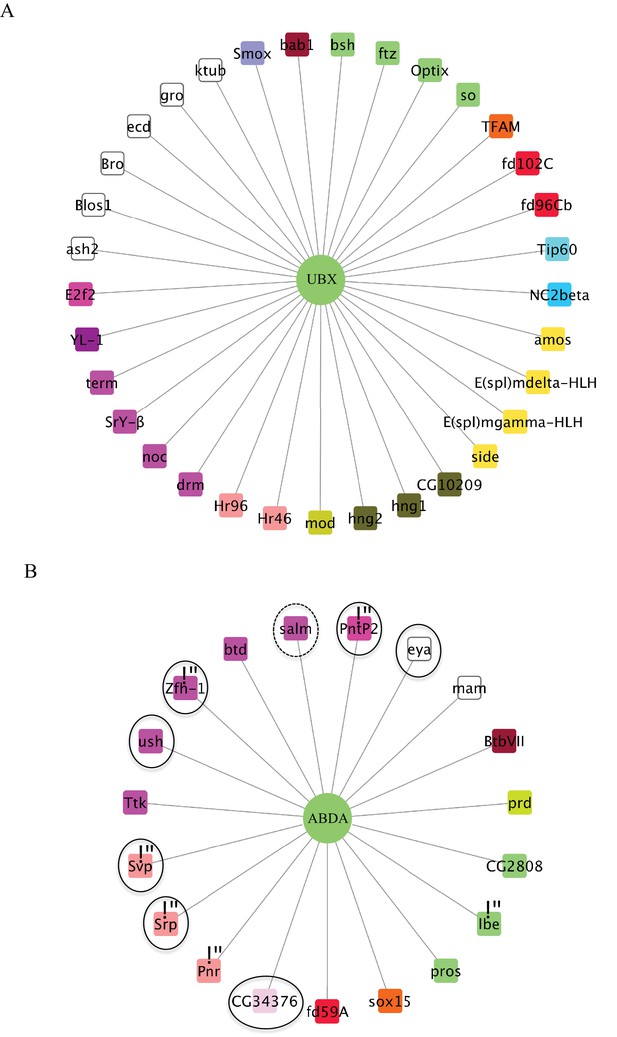
Representation of the Ubx- (A) and AbdA-specific (B) interactomes.
Colour code is as in Figure 3. TFs involved in gonad mesoderm or oenocytes specification, which correspond to AbdA-specific functions, are surrounded by a solid or dotted line, respectively. Star * denotes TFs involved in heart specification.
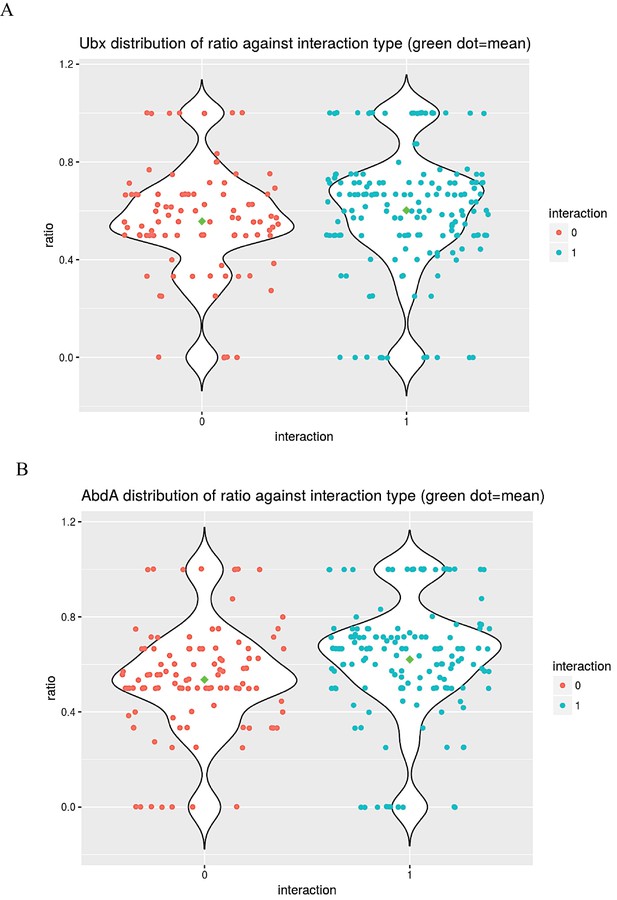
Analysis of the co-expression and interaction status of Ubx (A) or AbdA (B) and the positive ORFs.
The distributions of the values taken by the ratio between the number of tissues in which the TF and Ubx or AbdA are co-expressed and the total number of tissues composing the TF expression domain during embryogenesis (among the 25 analysed developmental contexts) with regard to their interaction status (red and blue dots indicate negative (0) and positive (1) interaction status between the TFs and Hox proteins, respectively). Green circles show the means of the distributions.
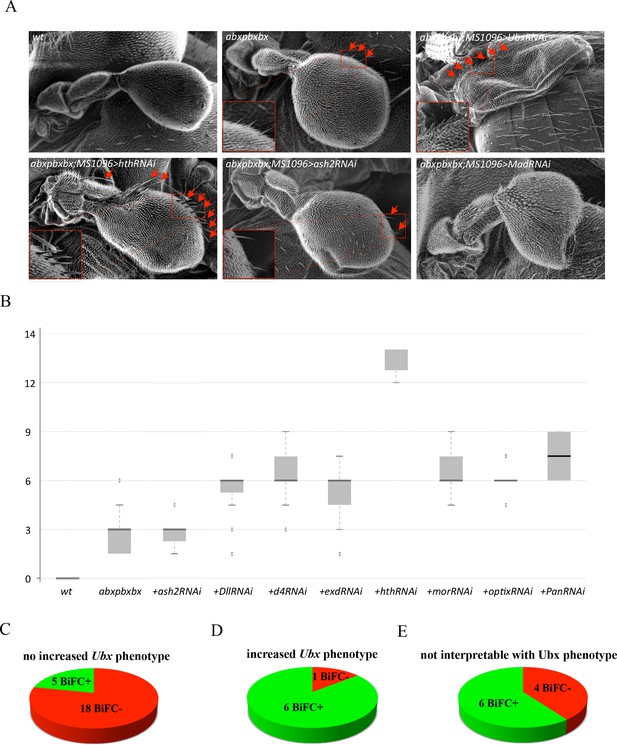
Functional genetics validates BiFC observations with Ubx in haltere primordium.
(A) Scanning electron microscopy of haltere phenotypes in the different genetic backgrounds, as indicated. Compared to wild-type, halteres of individuals heterozygous for the Hox regulatory mutation abxpbxbx have ectopic short wing-like hairs (arrows and zoom in/enlargement in B). This phenotype is increased when affecting Ubx expression upon expression of RNAi or when expressing a RNAi against a TF (although to a lesser extent: shown here for Homothorax, Hth) that could be required for Ubx function (arrows and enlargements). In contrast, expression of RNAi against a TF that is not required for Ubx function (as shown for absent_small_or_homeotic_discs _2, ash2) does not increase the phenotype. The expression of RNAi against TFs can also affect more globally the haltere (and wing) formation (as shown for Mad), which is difficult to interpret in term of homoeotic transformation and therefore with regard to a potential Ubx cofactor function. (B) Box plot statistical quantification of the haltere-to-wing transformations in the different genetic backgrounds, as indicated. Quantification was performed by counting the number of ectopic wing-like hairs formed at the edge of the haltere and on the hinge. The phenotype induced by the UbxRNAi was voluntary not included since it corresponds to an almost complete haltere-to-wing transformation. (C-E) Diagrams showing the distribution of TFs that were BiFC positive (green) and negative (red) with Ubx in the different cases (not increased haltere phenotype (C); increased haltere phenotype (D); not interpretable (E)). See also Supplementary file 6.
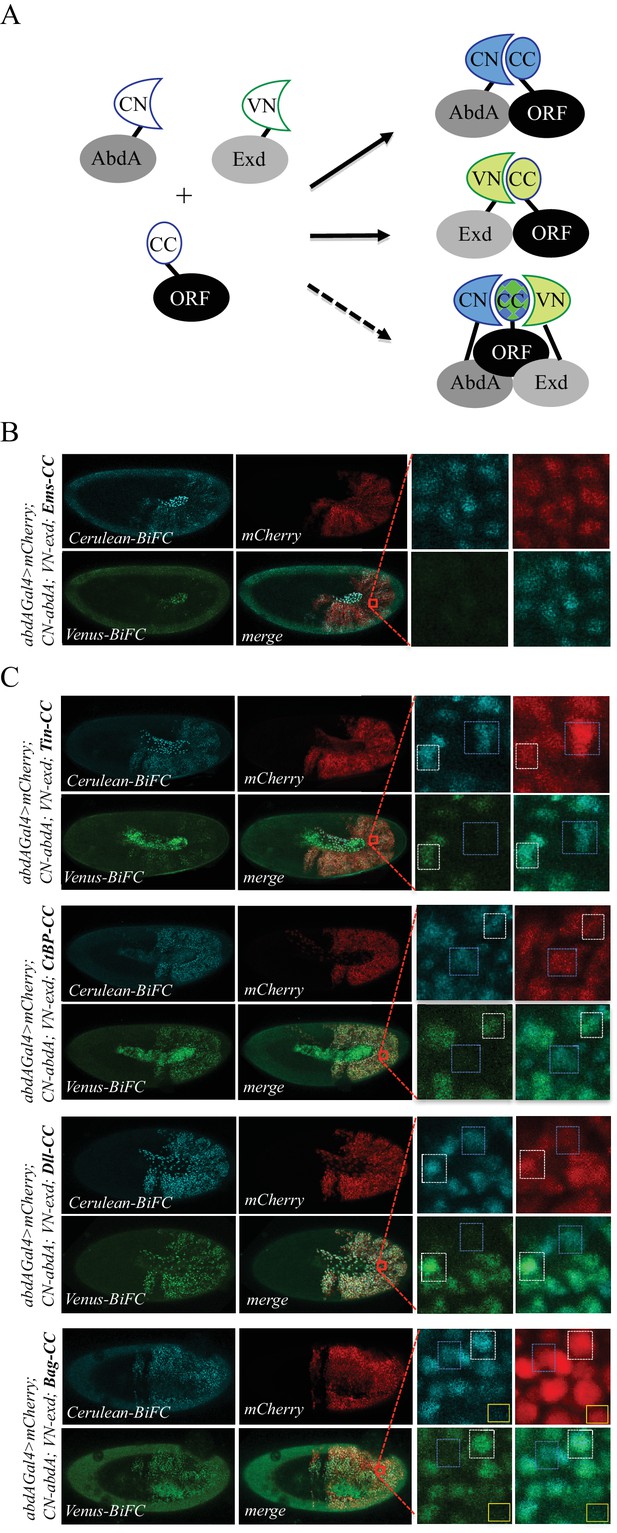
Using the multicolour BiFC library for analysing two different interactions in the same embryo.
(A) Principle of the bicolour BiFC. The AbdA and Extradenticle (Exd) cofactor are respectively fused to the CN or VN fragment, which can complement with the CC fragment of a co-expressed ORF when interaction occurs. The simultaneous expression of the three fusion proteins allows assessing Venus- and Cerulean-based BiFC in the same cell. Bicolour BiFC results from the interaction of the ORF-CC with both CN-AbdA and VN-Exd, thus revealing two binary interactions simultaneously. BiFC could result from interactions occurring in two independent complexes but potentially also in the context of a trimeric complex (dotted arrow) in vivo. (B) Illustrative confocal capture of stage 10 embryo expressing Empty spiracles (Ems) fused to CC, together with CN-AbdA and VN-Exd fusion proteins, as indicated. BiFC is only occuring between AbdA and Ems, as expected from previous observation. (C) Illustrative confocal captures of stage 10 embryos expressing CN-AbdA, VN-Exd and ORF-CC constructs, as indicated in the different panels (Tin: Tinman; CtBP; Distalless, Dll; Bagpipe, Bag). Enlargements are provided in each case. White-dotted boxes depict nuclei where the ORF-CC interacts with both AbdA and Exd. Blue-dotted boxes depict nuclei where the ORF-CC interacts only with AbdA. Yellow-dotted boxes depict nuclei with absence of interaction. Fusion proteins are under the control of the abdA-Gal4 driver. All expressing cells are recognized with the mCherry reporter. See also Figure 6—figure supplements 1–3 and Supplementary file 7.
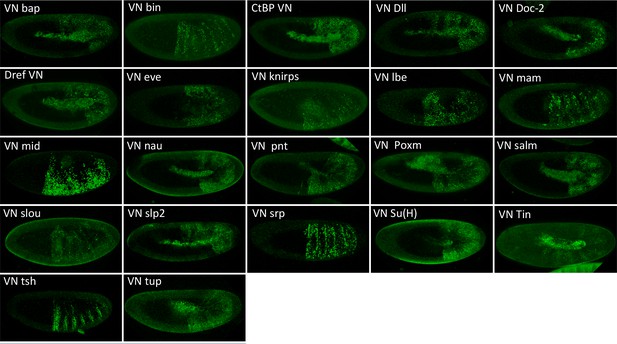
Illustrative confocal pictures of BiFC between ORF-VN and VC-Exd, as indicated.
Fusion proteins are expressed with the abdA-Gal4 driver and BiFC analysed in the epidermis of stage 10 embryos.
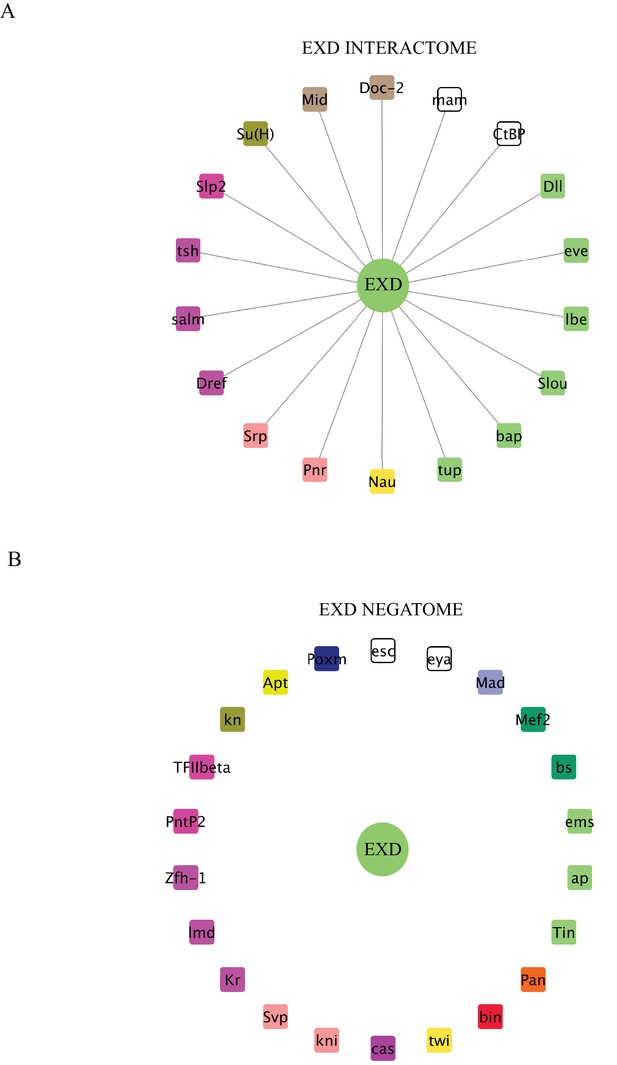
Interaction properties of Extradenticle (Exd) with a set of 37 TFs that are positive with AbdA.
(A) Representation of the interactome (positive interactions) of Exd. (B) Representation of the negatome (negative interactions) of Exd.
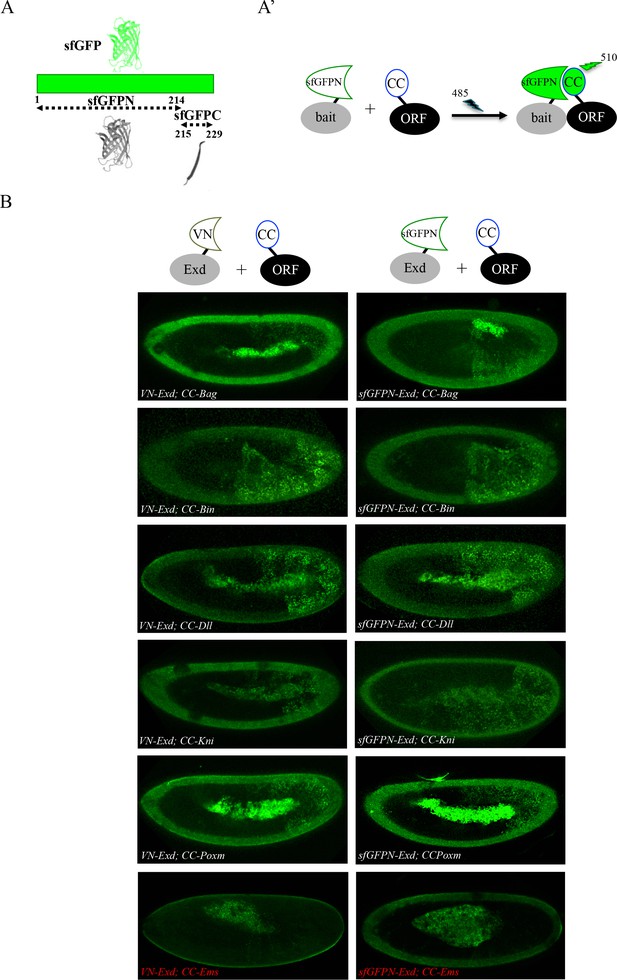
Adding the sfGFP to the fluorescence repertoire of the multicolour BiFC library.
(A) Split sfGFP fragments that are normally used for BiFC. (A’) Principle of BiFC with the N-terminal fragment of sfGFP (sfGFPN) and the C-terminal fragment of Cerulean (CC). Excitation and emission wavelengths of the sfGFP are indicated. (B) Illustrative confocal capture of BiFC obtained between ORF-CC constructs and VN-Exd (left panels) or sfGFPN-Exd (right panels), as indicated. Fusion proteins are expressed with the abdA-Gal4 driver and BiFC is analyzed in the epidermis of stage 10 embryos. Absence of interaction with Empty Spiracles (Ems) is highlighted in red.
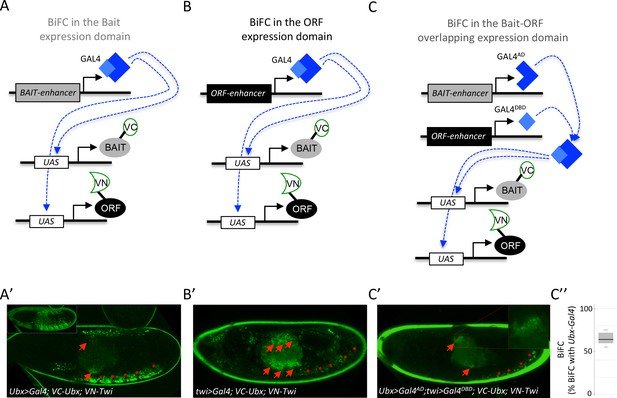
Coupling the multicolor BiFC library to the split-Gal4 system to visualize interactions in the overlapping expression domain of the two protein partners.
(A) Principle of BiFC with a unique Gal4 driver reproducing the expression profile of the bait protein (for example Hox-Gal4 driver). (B) Principle of BiFC with a Gal4 driver reproducing the expression profile of the ORF. (C) Principle of BiFC upon the independent expression of Gal4 moieties (Gal4AD and GALDBD) by using two different enhancers from the bait- or ORF-encoding gene. This system allows producing a functional Gal4 protein in the overlapping expression domain of the two enhancers, therefore assessing BiFC in cells that normally express both the bait and the ORF. (A’) Illustrative confocal picture of BiFC obtained upon the expression of Ultrabithorax (Ubx) and Twist (Twi) fusion proteins by using the Ubx-Gal4 driver. Confocal acquisitions were specifically obtained at the level of the visceral mesoderm to better highlight BiFC signals in the midgut (red arrow). Red stars depict signals in the somatic mesoderm. Insert shows acquisition of BiFC signals in the epidermis. (B') Illustrative confocal picture of BiFC obtained upon the expression of Ubx and Twi fusion proteins by using the twi-Gal4 driver. Confocal acquisitions were specifically obtained at the level of the visceral mesoderm. BiFC is occurring in the entire visceral mesoderm of the midgut (red arrows). Fluorescence is also occurring in cells of the somatic mesoderm (red stars). (C’) Illustrative confocal picture of BiFC obtained upon the expression of Ubx and Twi fusion proteins by using the split-Gal4 (Ubx-Gal4AD/twi-Gal4DBD) system. BiFC is occurring in the same specific part of the midgut as in A’ (enlargement on fluorescent nuclei is shown) and in few cells of the somatic mesoderm. C’’. Quantification of the fluorescence intensity obtained in the visceral mesoderm with the slipt-Gal4 system (using the fluorescence intensity obtained in the same tissue with the Ubx-Gal4 driver as a reference value). See also Supplementary file 9.
Additional files
-
Supplementary file 1
List of the 235 TFs of the multicolor BiFC library fused to the VN fragment at the C- or N-terminus, with a long or short linker region, as indicated.
Note that several TFs are fused to VN with either a long or a short linker region.
- https://doi.org/10.7554/eLife.38853.025
-
Supplementary file 2
List of the 326 TFs of the multicolor BiFC library fused to the CC fragment.
TFs highlighted in light green are also available as VN fusion constructs (see Supplementary file 1).
- https://doi.org/10.7554/eLife.38853.026
-
Supplementary file 3
List of the 260 VN- and CC-fusion TFs of the multicolor BiFC library that were tested with Ubx and AbdA.
- https://doi.org/10.7554/eLife.38853.027
-
Supplementary file 4
Expression profile of the 260 TFs that were tested with Ubx and AbdA among 25 different developmental contexts of the Drosophila embryo.
Each color code corresponds to a different tissue when expressed. Black boxes depict expression in tissues where Ubx and AbdA are not present. The last two columns indicate the number of co-occurrences of the TF and the Hox protein with regard to the total distribution (parentheses).
- https://doi.org/10.7554/eLife.38853.028
-
Supplementary file 5
Analysis of the proportion of common and specific AbdA- and Ubx interactors expressed in the different tissues, as indicated.
- https://doi.org/10.7554/eLife.38853.029
-
Supplementary file 6
List of TFs tested in RNAi in Ubx heterozygous mutant haltere discs.
Green and red boxes correspond to increased or not increased RNAi phenotypes, respectively. Black boxes correspond to RNAi phenotypes that could not be interpreted with regard to a potential Ubx cofactor function in the haltere disc (due to morphogenesis defects). Stars indicate RNAi fly lines that were tested with dicer. Stocks without the star are from the last generation and were tested without dicer.
- https://doi.org/10.7554/eLife.38853.030
-
Supplementary file 7
List of VN fusion TFs that were positive in BiFC tests with VC-AbdA and tested with VC-Exd.
Green and red boxes indicate a positive or negative interaction status, respectively.
- https://doi.org/10.7554/eLife.38853.031
-
Supplementary file 8
List of the 35 TFs tested as VN or CC fusion constructs with VC-AbdA or VN-AbdA, respectively.
Yellow boxes highlight the two TFs that showed opposite interaction status in the context of the two different fusion topologies (Hr83 and Ravus).
- https://doi.org/10.7554/eLife.38853.032
-
Supplementary file 9
Quantification of BiFC resulting from the overlap between twi- and Ubx-Gal4 drivers in the visceral mesoderm.
- https://doi.org/10.7554/eLife.38853.033
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38853.034





