A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis
Abstract
Morphogens provide positional information and their concentration is key to the organized development of multicellular organisms. Nitrogen-fixing root nodules are unique organs induced by Nod factor-producing bacteria. Localized production of Nod factors establishes a developmental field within the root where plant cells are reprogrammed to form infection threads and primordia. We found that regulation of Nod factor levels by Lotus japonicus is required for the formation of nitrogen-fixing organs, determining the fate of this induced developmental program. Our analysis of plant and bacterial mutants shows that a host chitinase modulates Nod factor levels possibly in a structure-dependent manner. In Lotus, this is required for maintaining Nod factor signalling in parallel with the elongation of infection threads within the nodule cortex, while root hair infection and primordia formation are not influenced. Our study shows that infected nodules require balanced levels of Nod factors for completing their transition to functional, nitrogen-fixing organs.
https://doi.org/10.7554/eLife.38874.001Introduction
Molecules of microbial origin have the capacity to induce a morphogenetic response in symbiotic eukaryotic hosts (Matsuo et al., 2005; Alegado et al., 2012; Lerouge et al., 1990). Nitrogen-fixing rhizobia produce species-specific decorated chitin-like molecules, Nod factors (Lerouge et al., 1990) that are recognized by LysM receptors in legume roots and initiate two different processes; nodule development and infection thread (IT) formation (Radutoiu et al., 2003; Murakami et al., 2018). In addition to their signalling capacity, Nod factors have been considered morphogens based on their effect on host developmental programs (Long, 1996), but direct measurements of in planta-produced Nod factor levels proved intractable, and therefore limited our knowledge on their concentration-dependent effect on the host (Morieri et al., 2013). ITs are tubular structures used by bacteria as conduits to advance from the root hair tip towards the root cortex. IT passage through cortical layers involves reprogramming of individual cells for microbial infection (van Brussel et al., 1992). Inside nodule primordia, branched IT arrays spread from the main shaft, guiding the confined rhizobia towards competent cortical cells where endocytosis takes place (Monahan-Giovanelli et al., 2006). Although a number of components are known (Xie et al., 2012; Yokota et al., 2009; Yano et al., 2009), the mechanisms governing IT formation are not defined, and even less understood is the control of IT branching inside root nodules. Rhizobia grow and multiply during infection thread progression (Fournier et al., 2008) and consequently bacterial signals accumulate inside the limited space within ITs. Nevertheless, the host determines the compatibility of Nod factors and exopolysaccharide produced by bacteria during root hair infection, via NFR1/NFR5 and EPR3 receptors, respectively (Kawaharada et al., 2015; Hayashi et al., 2014).
In parallel with the signalling events induced by Nod factor recognition, legumes express chitinases with Nod factor cleaving activity (Staehelin et al., 1994; Goormachtig et al., 1998). Plant chitinases are primarily found as components of immune responses induced by chitin producing pathogens (Lipka et al., 2005), but novel functions of these enzymes have evolved in plants (De Jong et al., 1992) and in symbiotic interactions between eukaryotes and microbes (Kremer et al., 2013). The identification of Nod factor-cleaving chitinases induced during legume-rhizobia symbiosis led to a model where these enzymes are required for Nod factor hydrolysis to avoid activation of immune responses induced by these chitin-like molecules during symbiosis (Goormachtig et al., 1998; Savoure et al., 1997; Staehelin et al., 2000). However, unlike chitin, Nod factors do not trigger immune responses in Lotus (Bozsoki et al., 2017) and in this context the function of Nod factor cleaving chitinases remained unclear. This was further confirmed by the Medicago truncatula, Nod factor hydrolase (NFH1) mutants that display delayed infection thread formation and nodule hypertrophy (Cai et al., 2018).
Results
We identified chit5-1, chit5-2 and chit5-3 as fix minus Gifu mutant alleles from screens aimed at discovering symbiotic defective mutations following inoculation with Mesorhizobium loti (Sandal et al., 2006) (Figure 1A). The impaired symbiotic association was reflected in significantly reduced shoot growth (Figure 1—figure supplement 1) and nitrogen deficiency symptoms when compared to wild type (Figure 1A). This defective phenotype was alleviated by the addition of nitrogen (10 mM KNO3) to the growth substrate (Figure 1—figure supplement 2). Detailed analysis of the symbiotic phenotype manifested by the three alleles revealed that nodule organogenesis was initiated in the presence of M. loti (Figure 1—source data 1), but most nodules remained arrested at the primordia stage with only a few developing into large, white or pink-spotted nodules (Figure 1B,C). Assessment of bacterial nitrogenase activity through acetylene reduction assays revealed that mutant nodules, despite their occasional functional appearance (pink-spotted), had significantly reduced nitrogenase activity compared to wild-type (Figure 1D). The limited nitrogen fixation capacity is reflected in the observed plant growth and nodule phenotypes. This was further supported by reduced transcriptional activation of leghemoglobin (Lb3) and sulfate transporter (Sst1) genes which are prominent molecular markers for mature nodules and successful nitrogen-fixing symbiosis (Ott et al., 2005; Krusell et al., 2005) (Figure 1—figure supplement 3). In contrast, transcripts associated with early rhizobial infection, early nodulin N6 (Mathis et al., 1999), pectate lyase Npl1 (Xie et al., 2012) and transcription factor Ern1 (Kawaharada et al., 2017a), were induced to higher levels in the chit5 mutants compared to wild-type plants (Figure 1—figure supplement 3). No morphological signatures associated with activation of plant defense responses, such as the onset of early senescence, previously reported for some fix minus mutants (Bourcy et al., 2013) was observed in chit5 plants (Figure 1B). Mpk3, Wrky29 and Wrky33, marker genes upregulated during plant immunity (Lohmann et al., 2010), were found to have a wild-type level of expression in both uninoculated and 21-dpi whole mutant roots (Figure 1—figure supplement 3 and Figure 1—source data 2). Recent transcriptome analyses of Lotus (Kelly et al., 2018a) revealed that progression of symbiosis in wild-type roots and formation of the symbiotic nodules (21 dpi) is accompanied by downregulation of these immunity marker genes (Figure 1—figure supplement 4A). We have analysed the expression of these marker genes in 21dpi nodules of wild-type and chit5 mutants. We found a higher level of the Npl1, Mpk3, Wrky29 and Wrky33 transcripts in chit5 mutant nodules compared to wild-type nodules (Figure 1—figure supplement 4). The differences in the expression levels of the early symbiotic and defense markers detected between wild-type and chit5 mutants indicate a deregulated symbiotic and immune signalling, seemingly arresting nodules at an early infection stage. We investigated whether rhizobial infection was affected and found that a normal number of ITs were formed in chit5 mutant root hairs (Figure 2A), and these proceeded in a manner similar to wild-type plants (Figure 2—figure supplement 1). Together, these results demonstrate that the early programs initiated by Nod factor signalling, nodule organogenesis and formation of root hair ITs, operate normally in chit5 mutants.
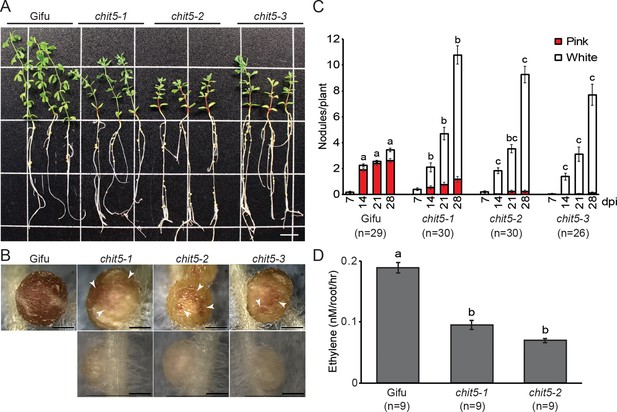
chit5 mutants are defective in nitrogen-fixing symbiosis.
(A) Three representative plants of wild-type Gifu and chit5 mutant alleles 6 weeks post-inoculation with M. loti R7A. Scale bar is 1 cm. (B) Gifu plants form pink nodules whilst chit5 mutants form mainly small white nodules, and occasionally pink-spotted (white arrows) nodules. Scale bars are 0.5 cm. (C) Average number of pink and white nodules formed by M. loti R7A over 4 weeks. (D) Nitrogenase activity measured as the amount of ethylene produced from acetylene in nodules of Gifu and chit5 mutants inoculated with M. loti R7A. (C) and (D) Error bars represent SEM and statistical comparisons of the number of pink nodules formed between genotypes at each time point are shown using ANOVA and Tukey post hoc testing with p values < 0.01 as indicated by different letters.
-
Figure 1—source data 1
Bacterial strains used in this study.
- https://doi.org/10.7554/eLife.38874.007
-
Figure 1—source data 2
Primers used for RT-qPCR analyses.
- https://doi.org/10.7554/eLife.38874.008
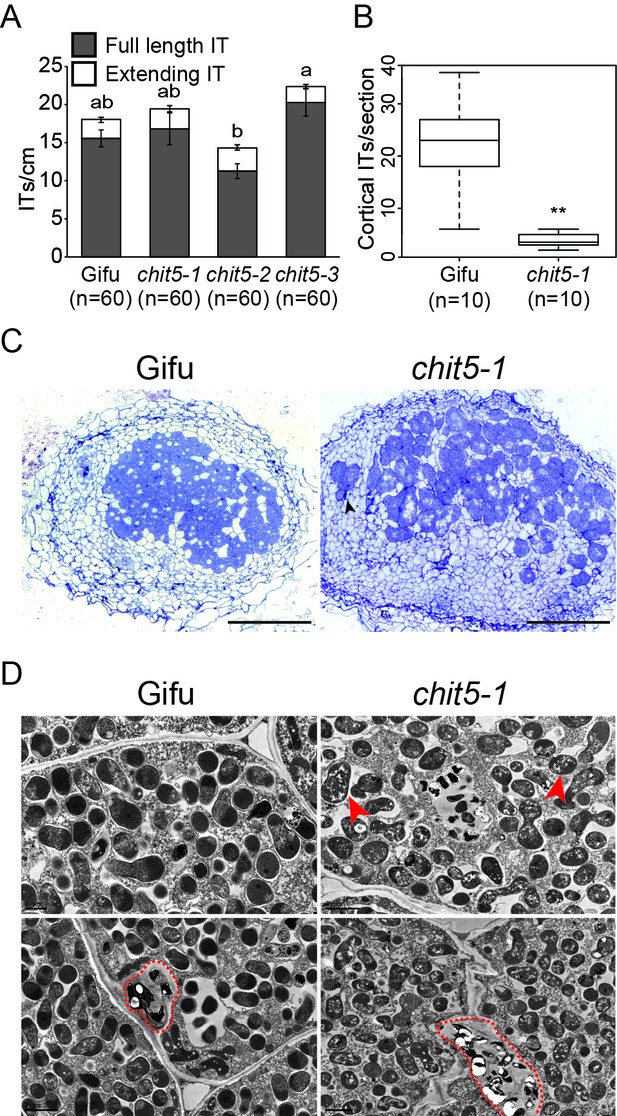
chit5 mutants have a defective nodule infection phenotype.
(A) Infection threads induced by M. loti + DsRed in Gifu and chit5 mutant root hair at 10 dpi . Error bars represent SEM and statistical comparisons of the number of full length infection threads formed between genotypes at each time point are shown using ANOVA and Tukey post hoc testing with p values < 0.01 as indicated by different letters. (B) Box plot of cortical infection thread counts performed on Gifu and chit5-1 nodule sections. t test p values are indicated by asterisks (**<0.01). (C) Light microscopy of nodule sections from the indicated plant genotypes 4 wpi stained with toluidine blue. Black arrows indicate bacteria between nodule cells. Scale bars are 200 µm (D) Transmission electron microscopy of nodule sections from the indicated plant genotypes 4 wpi. Red arrows point out bacteroids of stressed appearance with accumulated white spots, red dashed lines outline infection threads or invagination from the intercellular space. Note the difference between PHB bodies (large, white spots) characteristic for Mesorhizobium when present inside infection threads (red dashed outlines), and the small white spots present in the symbiosomes formed in the chit5 mutant nodules (red arrows). Scale bars are 2 µm. Corresponding images of chit5-2 and chit5-3 are in Figure 2—figure supplement 1.
In determinate nodules, like those formed by L. japonicus, ITs are present inside the cortex of mature nodules and can be quantified (Madsen et al., 2010). We have performed detailed investigations of IT phenotypes in wild-type and chit5 mutant nodule sections (n = 30) using light and electron microscopy. Counting of the cortical ITs formed inside wild-type and infected mutant nodules revealed a severe and significant reduction in their number in the mutant nodules (Figure 2B). Mutants displayed an impaired symbiotic occupancy of the nodule central tissue, with only scattered infected cells that formed inside the pink-spotted nodules (Figure 2C). Bacteria accumulated in the intercellular spaces between nodule cells (Figure 2C and D), giving rise to fewer, localized intracellular infections (Figure 2D). Symbiosomes present in the infected mutant cells contained multiple bacteroids of irregular shape and of atypical appearance (accumulation of white matrix) (Figure 2D). These phenotypes are likely a result of symbiotic and defense genes deregulation inside chit5 nodules. These results demonstrate that CHIT5 functions during cortical IT development and branching within nodules, and is required for the establishment of fully functional nitrogen-fixing symbiosis.
Map-based cloning and whole genome sequencing revealed that all three alleles are defective in one of the three chitinase-coding genes present at the identified genomic location (Figure 3A, Figure 3—figure supplement 1, and Figure 3—figure supplement 2), while the remaining two paralogs are pseudogenes with premature stop codons (Figure 3—figure supplement 2).The chit5-1 and chit5-3 mutants were found to have large genomic deletions encompassing the chitinase gene, while chit5-2 carries a point mutation (CCA to CTA), that results in a proline to leucine (P168-L) transition in the predicted protein sequence (Figure 3A and Figure 3—figure supplement 2). The symbiotic defective phenotype of all three alleles was restored by genetic complementation with the wild-type Chit5 gene expressed from its native promoter (2072 bp) and terminator (2108 bp) regions, or from the LjUbiquitin promoter and Nos terminator (Figure 3—figure supplement 3). Analyses of Chit5 expression using transcriptional reporters (Chit5-tYFPnls and Chit5-GUS) revealed promoter activity in all cells of uninoculated roots, in infected root hairs (Figure 3B), and nodule primordia (Figure 3—figure supplement 4). Chit5 promoter activity decreased inside fully functional nodules, indicating a reduced requirement for Chit5 at later stages of a functional symbiosis (Figure 3B, Figure 3—figure supplement 4).
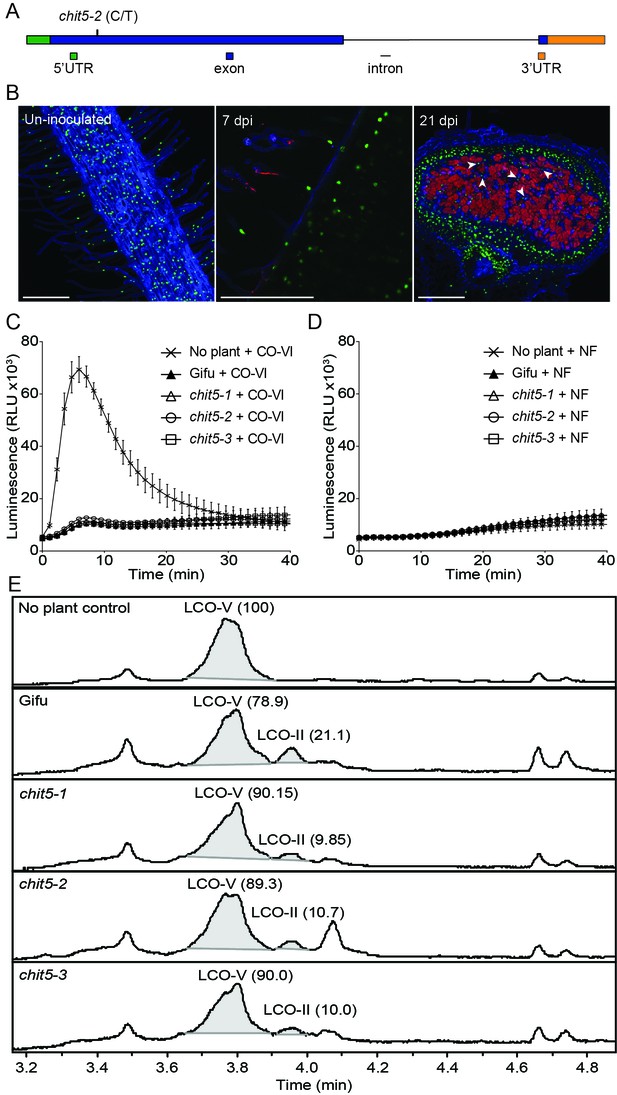
Chit5 encodes a root expressed class V chitinase with Nod factor hydrolase activity.
(A) Chit5 gene structure. Mutation in chit5-2 allele is shown. Chit5 gene is deleted in chit5-1 and chit5-3 alleles. (B) Chit5 promoter activity was monitored in Gifu roots transformed with a Chit5 promoter-tYFP-NLS (green nuclei) in uninoculated and M. loti R7A + DsRed inoculated roots. White arrows highlight examples of uninfected cells showing Chit5 promoter activity. Scale bars are 200 µm. (C) ROS induced by exudates of CO-VI-treated plants. (D) Absence of ROS induction by exudates of M. loti R7A Nod factor-treated plant exudates. (E) M. loti R7A Nod factor hydrolysis in the presence of the indicated plant genotypes measured by HPLC-MS. The average relative percentage of LCO-V and LCO-II fractions from two biological replicates (Figure 3—source data 1) determined from peak-peak integration is indicated in brackets. The peak eluting near 4.1 min is present in all plant treated samples but was not detected in the control sample. It lacks the characteristic fragmentation pattern of LCOs, and appears to be an aromatic, small molecule unrelated to chitinase activity.
-
Figure 3—source data 1
HPLC-MS analysis of Nod factor isolated after exposure to roots of wild-type Gifu or chit5 mutants.
- https://doi.org/10.7554/eLife.38874.017
The CHIT5 protein consists of a predicted signal peptide at the amino terminus followed by a class V (glycosyl hydrolase 18) domain with a conserved catalytic DxDxE motif (Figure 3—figure supplement 5). The proline residue mutated in chit5-2 allele is located immediately after the catalytic site, and is conserved across GH18-type chitinases, indicating a crucial role for the function of this protein (Figure 3—figure supplement 5). CHIT5 shares a high level of similarity to Medicago truncatula NFH1 (Zhang et al., 2016), including the presence of A and B loops and the proline residue (P294), previously reported as essential for Nod factor hydrolysis (Zhang et al., 2016). Furthermore, we found that a version of CHIT5 mutated in the DxDxE motif (E166 to K) was no longer able to complement the symbiotic defective phenotype, suggesting that enzymatic activity is required for CHIT5 function in planta (Figure 3—figure supplement 6). To investigate the enzymatic properties of CHIT5 in planta, we took advantage of its predicted secretion (Figure 3—figure supplement 5) and ubiquitous expression in Lotus roots (Figure 3B) and analysed the hydrolytic capacities of wild-type and mutant root exudates on chitin hexamers (hexa-N-acetylchitohexaose, abbreviated CO-VI) and on R7A Nod factor (NodMI-V(C18:1, Me, Cb, FucAc), abbreviated LCO-V).
For testing CHIT5-dependent hydrolysis of CO-VI by root exudates we made use of a physiological test based on the differential capacity of CO-VI, CO-III/CO-II and Nod factor elicitors to induce ROS production in Lotus (Bozsoki et al., 2017). CO-VI elicitors present in the control samples (not exposed to plant root exudates) induced ROS production, while those exposed to wild-type or mutant roots lost this capacity (Figure 3C). This shows that CO-VI hydrolysis took place during incubation with plant roots irrespective of the genotype and that CHIT5 is thus not solely required for CO-VI hydrolysis in the Lotus rhizosphere. Similar analyses performed with LCO-V showed no ROS production, confirming that intact R7A Nod factor molecules (present in the control samples) or their possible hydrolysis products resulting from exposure to plant roots, lack the ability to induce ROS in Lotus roots (Figure 3D) (Bozsoki et al., 2017). Analyses of butanol-extracted fractions from LCO-V samples determined that hydrolysis of the R7A Nod factor into NodMI-II (C18:1, Me, Cb, abbreviated LCO-II) was induced by exudates from roots (Figure 3E). Exudates from all chit5 mutants induced lower production of LCO-II than wild-type exudates, and there was a corresponding reduction in LCO-V degradation by mutants compared with wild-type (Figure 3E and Figure 3—source data 1). Since LCO-II was produced after exposure to chit5 exudates, additional plant chitinases expressed in Lotus roots (Figure 3—figure supplement 7) may contribute to Nod factor hydrolysis in the Lotus rhizosphere. Our in planta assays demonstrate that secreted CHIT5 contributes to hydrolysis of R7A Nod factor. Nevertheless, CHIT5 and unknown hydrolase(s) induce only a limited hydrolysis of the fully compatible Nod factor, most likely to ensure that symbiotic signalling occurs, as shown in wild-type plants.
M. loti NodD1 and NodD2 transcriptional regulators control the expression of Nod factor biosynthesis genes and are preferentially active at distinct stages of M. loti-Lotus symbiosis (Kelly et al., 2018b). NodD1 is primarily active inside root hair ITs, while NodD2 activates the transcription of Nod factor biosynthesis genes within nodules. NodD1 and NodD2 were found to differ significantly in their capacity to induce Nod factor biosynthesis genes transcription on Lotus roots and coordinated activity between them ensures an intermediate gene expression in wild-type R7A (Kelly et al., 2018b). Our analysis of chit5 mutants revealed a requirement of CHIT5 for cortical IT extension leading to effective colonization of nitrogen-fixing nodules (Figure 2). This provided us with the opportunity to further investigate the necessity of CHIT5 for monitoring Nod factor levels in planta, by assessing the phenotypes of nodD1 (D1-/D2+) and nodD2 (D1+/D2-) bacterial mutants. chit5 mutants inoculated with nodD1, that induces a 3-fold higher level of Nod factor biosynthesis gene transcription compared to wild-type R7A in response to Lotus root exudates (Kelly et al., 2018b), displayed a severe nitrogen-deficient phenotype (Figure 4A). Comparable root hair IT formation was observed on wild-type and chit5 plants (Figure 4—figure supplement 1) yet nodules induced by nodD1 on chit5 mutants were even more severely impaired than those induced by R7A. Nodules were rarely or superficially infected and were found to be severely ineffective in acetylene reduction assays (Figure 4 and Figure 4—figure supplement 1). In contrast, inoculation with nodD2, that induces a 4-fold lower level of Nod factor biosynthesis gene transcription compared to wild-type R7A (Kelly et al., 2018b), resulted in full restoration of chit5 defective phenotypes (Figure 4 and Figure 4—figure supplement 1). A similar number of pink nodules were formed on wild-type and chit5 mutant roots and plants were no longer nitrogen-deficient. Fully colonized nodules with efficient nitrogen-fixing symbiosomes were formed (Figure 4—figure supplement 1). These contrasting phenotypes of chit5 mutants in response to M. loti strains affected in the regulation of Nod factor biosynthesis indicate that CHIT5-dependent modulation of Nod factor levels within nodules is critical for cortical IT development and complete transition to nitrogen-fixing symbiosis.
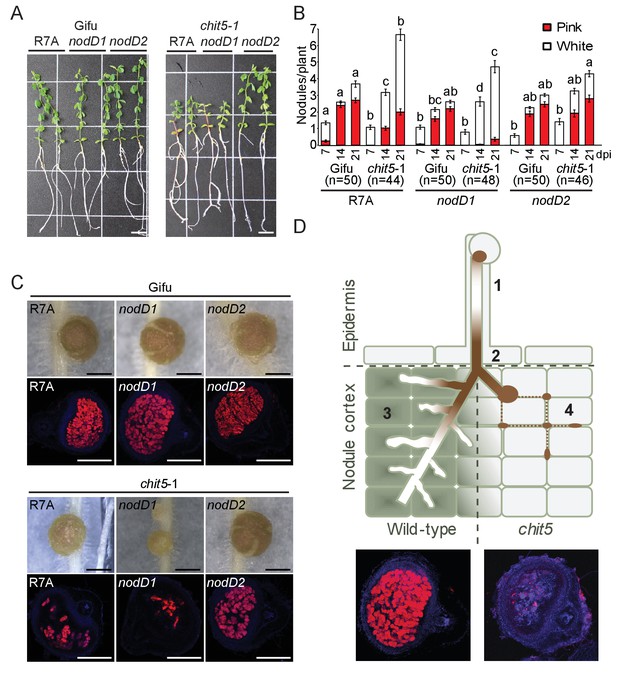
The symbiotic impairment of chit5 mutants can be overcome by an M.loti R7A mutant affected in the regulation of Nod factor biosynthesis.
(A) Representative images of Gifu and chit5-1 plants inoculated with the indicated M. loti R7A strains 7 wpi. Scale bars are 1 cm. (B) Average number of pink and white nodules formed on Gifu and chit5-1 by the indicated M. loti R7A strains over 3 weeks. Error bars represent SEM and statistical comparisons of the number of pink nodules formed between strains on each genotype at each time point are shown using ANOVA and Tukey post hoc testing with p values < 0.05 as indicated by different letters. (C) Light microscopy of whole nodules 3 wpi and confocal images of nodule sections from Gifu and chit5-1 plants inoculated with the indicated strains tagged with DsRed. Scale bars are 0.5 cm. (D) Proposed model of CHIT5 activity and representative nodule images of wild-type and chit5 mutant illustrating the observed phenotype. (1) Bacteria maintain a low level of Nod factors within root hair-traversing ITs by preferential activity of NodD1. (2) and (3) Bacterial amplification inside ITs, coupled with the switch for preferential activity of NodD2 leads to higher levels of fully decorated pentameric Nod factors (LCO-V). Chit5 expression (green filled cells in wild-type) is crucial for maintaining a balanced Nod factor level enabling IT extension inside primordia, efficient bacterial endocytosis and, ultimately, development of the nitrogen-fixing organ (3). (4) In the chit5 mutants (light grey filled cells) higher levels of Nod factors (LCO-V) impede IT elongation and branching inside primordia leading to bacteria accumulation in between the cells (dotted brown line) and scattered infection.
Nod factors are perceived by the NFR1/NFR5 receptors (Radutoiu et al., 2003). The expression of Chit5 is similar to that of Nfr1/Nfr5 (Kawaharada et al., 2017b) and follows the spatio-temporal development of symbiosis in the root and nodules (Figure 3B and Figure 3—figure supplement 4). The observed requirement for tight modulation of Nod factor levels inside nodules (Figure 4A and Figure 4—figure supplement 1) prompted us to investigate whether the chit5 phenotype in the presence of wild-type M. loti could reflect a bottleneck in Nod factor signalling due to limited/tightly controlled NFR-dependent signalling. We tested this possibility by overexpressing Nfr1 and Nfr5 receptors in chit5 mutants, but observed no change in their symbiotic phenotype after inoculation with M. loti R7A (Figure 4—figure supplement 2). This indicates that increased and ectopic expression of Nod factor receptors is insufficient to restore the chit5 defective phenotype.
Bacterial genetics coupled with in planta phenotypic studies and chemical analyses of Nod factors produced by rhizobial mutants have determined that various Nod factor decorations are required for initiation and progression of symbiosis (Ehrhardt et al., 1995; Rodpothong et al., 2009). We investigated the importance of Nod factor decorations for CHIT5 activity in Lotus by assessing the phenotypes induced by R7A nolL and nodZ mutants that produce Nod factors lacking acetyl (LC(C18:1, Me, Cb, Fuc)) and acetylated-fucosyl decorations (LCO-V(18:1, Me, Cb)), respectively on the reducing-end of the Nod factor (Rodpothong et al., 2009). Compared to wild-type M. loti, these bacterial mutants are delayed in initiating nodule organogenesis and form fewer infected nodules (Figure 4—figure supplement 3), a phenotype similar to the one induced by M. loti nodD1 mutant (Kelly et al., 2018b). Interestingly, we found that these bacterial mutants infected chit5 mutants and wild-type similarly and that, compared to wild-type or M. loti nodD1, no significant differences in nodule number, infection rate and nitrogen fixing ability were observed between plant genotypes (Figure 4—figure supplement 3). This suggests that cortical infection and onset of nitrogen fixation in the symbiotic organ might require tight modulation of Nod factor levels in a structure-dependent manner. Decorations have been shown to be important for determining the rate of Nod factor degradation by different chitinases (Ovtsyna et al., 2000). Nod factors produced by nodZ and nolL mutants may become accessible to other chitinases present in the chit5 mutants for degradation. Alternatively, Nod factor signalling induced by nodZ and nolL mutants may be reduced compared to wild-type bacteria and therefore not requiring CHIT5-dependent modulation. These findings based on analyses of plant and bacterial mutants in binary interactions questioned the functional relevance of CHIT5 in the context of natural environments where a diverse rhizobial population, presumably producing Nod factors with various structures at various levels, is present. We investigated this by analysing the phenotypes of chit5 mutant plants grown in soil and exposed to the natural rhizobial population compared with wild-type and nfr5-2 plants. We found that both chit5 and nfr5 mutant plants were clearly distinctive from wild-type, as they were nitrogen starved in spite of numerous nodule primordia formed on chit5 plants (Figure 4—figure supplement 4). This indicates, that at least in the presence of the tested soil rhizobial population, CHIT5 is a major determinant for the onset of nitrogen-fixing symbiosis in the nodules whose formation is controlled by NFR5.
Discussion
Our study revealed that in L. japonicus, CHIT5-dependent hydrolysis of the Nod factor morphogen plays a crucial role during primordia infection and for full-transition to symbiotic nitrogen fixation. Chit5 in L. japonicus and Nfh1 in M. truncatula are highly similar, however the mutant phenotypes in the two model-legumes are very different (Cai et al., 2018). Unlike in the nfh1 mutant (Cai et al., 2018), the number and appearance of root hair ITs and the size of nodule primordia appear not to be affected in chit5 mutants, indicating that in Lotus other mechanisms independent of CHIT5 control these early stages of symbiosis. Chit5 and Nfh1 are part of genomic clusters with closely-located paralogs (Figure 3—figure supplement 1, Figure 4—figure supplements 5 and 6). In Lotus, two of the three paralogs evolved as pseudogenes (Figure 3—figure supplement 2 and Figure 4—figure supplement 7), while in Medicago all are functional genes and expressed (Figure 4—figure supplement 5). This may contribute to the differential phenotypes observed in the two model legumes, and may explain the strong phenotype displayed by Chit5 mutants in Lotus. Alternatively, Chit5 and Nfh1 might be highly similar but non-orthologous genes. Based on our observations from Lotus we propose a likely scenario explaining CHIT5 function (Figure 4D). Bacteria maintain a low level of Nod factors within root hair-traversing ITs by preferential activity of NodD1 (Kelly et al., 2018b). Bacterial amplification inside primordia-elongating ITs, coupled with the switch for preferential activity of NodD2 (Kelly et al., 2018b) leads to higher levels of fully decorated pentameric Nod factors (LCO-V) that unbalance the symbiotic and immune signalling in the cortical cells. CHIT5 hydrolytic activity on LCO-V is crucial for maintaining balanced symbiotic and defense signalling in the root developmental field and is required for cortical IT extension inside primordia, efficient bacterial endocytosis and ultimately, development of the nitrogen-fixing organ. In the absence of Chit5, IT elongation is impaired leading to bacteria accumulation in the intercellular space and sparse intracellular infection (Figure 4D). However, we cannot exclude that a CHIT5-dependent product derived from Nod-factor hydrolysis plays a role during nodule infection. Interestingly, similar but less frequent infections, as observed in chit5 mutants inoculated with wild-type M. loti, was reported for spontaneous nodules formed on nfr1snf1 plants when infected by an M. loti nodC mutant lacking the ability to produce Nod factors. Surprisingly, the symbiosomes formed by the M. loti nodC mutant were reported to have normal appearance and the plants to be nitrogen proficient (Madsen et al., 2010). These results suggest that Nod factors, or their hydrolytic products, are dispensable for nitrogen fixation within infected cells. Based on these results and our findings from symbiotic gene expression in chit5 mutants and their phenotype in the presence of mutant rhizobia, we consider the scenario proposed in Figure 4D as the most likely framework for CHIT5 function. Future studies will likely reveal which molecular determinants are responsible for decoding the CHIT5 output into nitrogen-fixing state inside infected cells.
Materials and methods
Plant materials
Request a detailed protocolLotus japonicus ecotype Gifu B-129 (Handberg and Stougaard, 1992) was used as the wild-type plant. Plants were grown at 21°C with 16 h day and 8 hr night cycles. Agrobacterium strain AR1193 (Stougaard et al., 1987) was used for hairy-root transformation experiments, carried out as described previously (Petit et al., 1987). Plants were inoculated with corresponding bacterial suspensions, OD600 = 0.02.
Bacterial strains and plasmids
Request a detailed protocolBacterial strains and plasmids used in this study are listed in Figure 1—source data 1. Mesorhizobium loti R7A (Sullivan et al., 2002; Kelly et al., 2014) and mutant strains were cultured at 28°C in YMB or G/RDM medium (Ronson et al., 1987). Strains expressing fluorescent reporters were constructed by the introduction of pSKDSRED or pSKGFP (Kelly et al., 2013). Antibiotics were added to media as required at the following concentrations: tetracycline, 2 μg ml−1; rifampicin, 100 μg ml−1; spectinomycin, 100 μg ml−1; ampicillin, 100 µg ml−1.
Chit5 gene cloning
Request a detailed protocolThe chit5-1 (sym43), chit5-2 (sym103-1) and chit5-3 (sym103-2) mutants were isolated as fix- mutants in the background of L. japonicus ecotype ‘Gifu’, and they were described by Sandal et al. (2006). An F2 mapping population was established by crossing chit5-1 mutant to wild-type L. japonicus ecotype ‘MG-20’. In total, 1508 F2 offspring mutant plants allowed us to limit the region of interest to 483 kb on chromosome 5 (Figure 3—figure supplement 1). Additional attempts to further limit the candidate region were not successful due to suppression of recombination in this chromosomal region. Next, we used a combination of BAC/TAC subcloning and whole genome sequencing of nuclear DNA (Schneeberger et al., 2009) from the three alleles by Illumina technology (NexTera Library Kit) to pinpoint the candidate mutated gene. SHOREmap strategy (Schneeberger et al., 2009) was used for SNP calling based on L. japonicus v.3.0 genome. We identified that chit5-1 and chit5-3 contain deletions of app. 13 and 9 kb, respectively in this region while chit5-2 has a point mutation C/T in a predicted chitinase gene. Three chitinases (Chit5, Chit5a and Chit5b) sharing high identity in the coding, 5’ and 3’ DNA flanking regions were identified to be present in this genomic region (Figure 3—figure supplement 2). Chit5a and Chit5b have early stop codons and are therefore pseudogenes (Figure 3—figure supplement 2). The chit5-1 and chit5-3 mutants lack Chit5 gene, and one of the pseudogenes, while chit5-2 has all three genes, but contains a point mutation (C/T) in Chit5. The precise location of the three genes in the current version of L. japonicus genome is not possible to assign due to the high level of similarity and repetitive nature of the DNA sequences present in this region.
Plant phenotypic assessment
Request a detailed protocolSeed sterilization and plant-growth setups for nodulation and infection thread assays were as previously described (Kawaharada et al., 2015). For nodulation assays plants were inoculated with R7A, nodD1, nodD2, nolL and nodZ and scored weekly. For IT counts, plants were inoculated with strains carrying the pSKDSRED reporter plasmid. Roots of 10 dpi- plants (n = 20 per biological replica) were cut into 1 cm pieces and 20 pieces were examined for infection thread counts. For phenotypic assessment of plants grown in soil, sterile seedlings were planted in Cologne soil (Zgadzaj et al., 2016) and the shoot biomass, together with the number of pink and total nodules were counted at 9 weeks post-planting. The soil was supplemented with 10 mM KNO3 for phenotypic assessment in the presence of nitrogen.
Acetylene reduction assays
Request a detailed protocolEthylene production was monitored in acetylene reduction assays using a SensorSense (Nijmegen, NL) ETD-300 ethylene detector as previously described (Reid et al., 2018).
RT-qPCR
Request a detailed protocolRoot systems formed by chit5 mutants and wild type plants were harvested 21 days after inoculation with M. loti R7A mock (diluted YMB medium). The plants were grown on plates supplemented with ¼ B and D medium in a 16/8 hr day/night regime of 21/16°C. The mRNA was extracted using Dynabeads (Invitrogen) and the quality of the purified mRNA was examined with a 2100 Bioanalyzer RNA pico chip (Agilent). cDNA was synthesized by RevertAid Reverse Transcriptase (Fermentas) with an OligodT primer. For quantitative real-time PCR analysis of symbiotic transcripts levels, three housekeeping genes (ATP, UBC, and PP2A encoding L. japonicus ATP synthase, Ubiquitin Conjugating Enzyme and Protein Phosphatase 2A) were used as references. Specificity of the primers was ensured by melting curve analysis and sequencing of the amplification products. Quantitative real time PCR using specific primers for the reference and target genes (Figure 1—source data 2) was performed using the LightCycler 480 II (Roche) with the LightCycler SYBR Green I Master kit. The relative quantification software by Roche was used to determine the efficiency-corrected relative transcript levels, normalized to a calibrator sample. The geometric mean of the relative expression ratios for the three biological and three technical replicates has been calculated (Vandesompele et al., 2002) as well as the corresponding upper and lower 95% confidence intervals.
Light microscopy (LM) and transmission electron microscopy (TEM) analyses including cortical infection thread (IT) counts.
Request a detailed protocolFor cortical ITs white nodules inoculated with ML001 DsRed were scored at four wpi. Nodules were prepared for LM and TEM investigation of infection from the indicated genotypes. Nodules that were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate (pH 7.0) overnight at 4°C were dehydrated in an ethanol series, and embedded in LR White acrylic resin (Agar Scientific, UK). Semithin sections (1 μm) of these were taken for light microscopy. Ultrathin (70 nm) sections were taken for TEM using a Leica UCT ultramicrotome from samples that had been post-fixed in 1% osmium tetroxide and embedded in Araldite epoxy resin. The semithin sections were collected on glass slides and stained with 0.1% toluidine blue, whereas the ultrathin sections were collected on pioloform-coated nickel grids. The number of ITs in semithin sections was counted in two representative sections of 10 different nodules for wild-type and chit5-1 genotypes.
Chit5 expression analyses
Request a detailed protocolChit5 promoter region of 2072 bp upstream of start codon was PCR-amplified from L. japonicus Gifu using primers caccCATACTTAACCAATGTGGTACTTCAATTC (PM-9331) and GTGTATATATATGTGAAACCTTGCATCTC (PM-9332). The amplification product was cloned into pIV10 carrying GUS or tYFP-NLS (Reid et al., 2017) reporters using a Gateway cloning strategy. Promoter activity was investigated in transformed roots expressing the reporter constructs at 1, 3, 7, 10, 14, 21 dpi after inoculation with M. loti. For analysis of Chit5 promoter-GUS activity, roots were stained as previously described (Kawaharada et al., 2017a). Chit5 promoter-tYFP-NLS activity was investigated using a Zeiss LSM780 meta confocal microscope.
Genetic complementation
Request a detailed protocolFor complementation analysis, a construct consisting of Chit5 promoter (2072 bp) followed by genomic Chit5 sequence (3206 bp) was used. The clone was obtained by a sequential subcloning from BAC69G19 following: 1) HindIII digest and cloning in pGreen29, and 2) SmaI and SalI digest of pGreen29 construct, and cloning into pIV10. The construct was integrated into Agrobacterium rhizogenes AR1193 that was used for hairy root plant transformations. AR1193 with empty pIV10 vector was used as a control. To introduce the E166 to K mutation in CHIT5 sequence, the piV10 Chit5 construct was used as a template for introducing the point mutation using primers GATTGGaAGTGGCCAGGAGATG (PM-11341) and CCACTtCCAATCCAAGTCAAGACC (PM-11342). The construct was then moved to Agrobacterium rhizogenes AR1193 for hairy root plant transformations. For Chit5 overexpression the Chit5 coding region from start to stop codon was amplified using pIV10-Chit5 as template and primers caccATGATCATCAAGCTCTTGGTTGC (PM-9870) and TCAATCATTATAAAGAGGTGAAAACAAGTG (PM-9829), followed by cloning into pIV10 vector containing the LjUbiquitin promoter and Nos terminator using Gateway cloning strategy. For Nfr1/Nfr5 over-expression the constructs described by Radutoiu et al. (Radutoiu et al., 2007) have been used. All constructs have been sequence-verified.
Root exudate hydrolysis assays
Request a detailed protocolThe assays were performed as described by Staehelin et al (Staehelin et al., 1995). with adaptation for L. japonicus. Surface sterilized seeds of wild-type and chit5 mutants were germinated on upright plates with wet filter paper in a 16/8 hr day/night regime of 21/16°C for 3 days. The emerging plant roots were pre-treated with R7A Nod factor by transfer into 0.5 ml dark glass vials and grown O/N in ¼ B and D medium supplemented with 0.1 μM NodMI-V(C18:1, Me, Cb, FucAc). Pre-treated seedlings were transferred to new 0.5 ml dark glass vials filled with 300 μl of ¼ B and D medium supplemented with 10 μM NodMI-V(C18:1, Me, Cb, FucAc) or hexa-N-acetylchitohexaose (CO- VI) for 18 hr incubation time in the dark. Seedlings were removed and the incubating solution was used for measurements of ROS induction as described by Bozsoki et al. (Bozsoki et al., 2017). Nod factor hydrolysis by root exudates was determined by extracting the Nod factors from the incubating solution with equal volume of distilled n-butanol for extraction. Freeze dried samples were analysed as described below.
Analyses of Nod factor hydrolysis
Request a detailed protocolThe n-Butanol-extracted samples from the root exudate hydrolysis assay were dissolved in acetonitrile-water mixture (1:1 (v/v), 50 μL) and analysed on a Dionex UltiMate 3000 UHPLC+ focused system. Separation of LCOs was performed using a C4 column (Phenomenex Aeris 3.6 µm widepore, 200 Å, 50 × 2.1 mm). Samples were eluted by applying a gradient of CH3CN in H2O containing 0.1% formic acid with a flow rate of 1 mL/min for 12 min. Peak integration was performed using the wavelength range 190–440 nm to maximize signal intensity. HPLC data were complemented by high-resolution mass spectrometric identification of LCO-V (NodMl-V(C18:1, Me, Cb, FucAc), and hydrolysis products LCO-III (NodMl-III(C18:1, Me, Cb)) and LCO-II (NodMl-II(C18:1, Me, Cb)) using a Bruker Impact HD UHR-QTOF mass spectrometer connected to the LC. HR-MS (ESI-TOF): calcd. for LCO-V, [M + H]+: m/z 1501.7394; found 1501.7414. HR-MS (ESI-TOF): calcd. for LCO-II, [M + H]+: m/z 704.4328; found 704.4346.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
-
A carrot somatic embryo mutant is rescued by chitinaseThe Plant Cell Online 4:425–433.https://doi.org/10.1105/tpc.4.4.425
-
The ethylene responsive factor required for nodulation 1 (ERN1) Transcription factor is required for Infection-Thread formation in Lotus japonicusMolecular Plant-Microbe Interactions 30:194–204.https://doi.org/10.1094/MPMI-11-16-0237-R
-
Conditional requirement for exopolysaccharide in the Mesorhizobium-Lotus symbiosisMolecular Plant-Microbe Interactions 26:319–329.https://doi.org/10.1094/MPMI-09-12-0227-R
-
Genome sequence of the Lotus spp. microsymbiont Mesorhizobium loti strain R7AStandards in Genomic Sciences 9:.https://doi.org/10.1186/1944-3277-9-6
-
Evolution and regulation of the Lotus japonicus LysM receptor gene familyMolecular Plant-Microbe Interactions 23:510–521.https://doi.org/10.1094/MPMI-23-4-0510
-
Rhizobium symbiosis: nod factors in perspectiveThe Plant Cell Online 8:1885–1898.https://doi.org/10.1105/tpc.8.10.1885
-
The early nodulin gene MtN6 is a novel marker for events preceding infection of Medicago truncatula roots by Sinorhizobium melilotiMolecular Plant-Microbe Interactions 12:544–555.https://doi.org/10.1094/MPMI.1999.12.6.544
-
Transformation and regeneration of the legume Lotus corniculatus: a system for molecular studies of symbiotic nitrogen fixationMGG Molecular & General Genetics 207:245–250.https://doi.org/10.1007/BF00331585
-
Dynamics of ethylene production in response to compatible nod factorPlant Physiology 176:1764–1772.https://doi.org/10.1104/pp.17.01371
-
Nodulation gene mutants of Mesorhizobium loti R7A-nodZ and nolL mutants have host-specific phenotypes on Lotus sppMolecular Plant-Microbe Interactions 22:1546–1554.https://doi.org/10.1094/MPMI-22-12-1546
-
Rhizobium meliloti ntrA (rpoN) gene is required for diverse metabolic functionsJournal of Bacteriology 169:2424–2431.https://doi.org/10.1128/jb.169.6.2424-2431.1987
-
Genetics of symbiosis in Lotus japonicus: recombinant inbred lines, comparative genetic maps, and map position of 35 symbiotic lociMolecular Plant-Microbe Interactions 19:80–91.https://doi.org/10.1094/MPMI-19-0080
-
The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plantsMGG Molecular & General Genetics 207:251–255.https://doi.org/10.1007/BF00331586
-
Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7AJournal of Bacteriology 184:3086–3095.https://doi.org/10.1128/JB.184.11.3086-3095.2002
Article and author information
Author details
Funding
Danish National Research Foundation (DNRF79)
- Anna Malolepszy
- Simon Kelly
- Kasper Kildegaard Sørensen
- Christina Kalisch
- Zoltan Bozsoki
- Michael Panting
- Stig U Andersen
- Dorthe Bødker Jensen
- Maria Vinther
- Noor de Jong
- Lene Heegaard Madsen
- Kira Gysel
- Mette U Berentsen
- Mickael Blaise
- Knud Jørgen Jensen
- Mikkel B Thygesen
- Niels Sandal
- Kasper Røjkjær Andersen
- Simona Radutoiu
China Scholarship Council (201604910506)
- Ke Tao
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Clive Ronson and John Sullivan for providing the M. loti R7A mutants. We thank Keisuke Yokota for his contribution to fine mapping of Chit5. We thank Finn Pedersen for plant care. We thank Clive Ronson and Christian Staehelin for constructive discussions. We thank Jens Stougaard for critical reading of the manuscript. This work was supported by the Danish Research Foundation, DNRF 79 grant.
Copyright
© 2018, Malolepszy et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 3,027
- views
-
- 633
- downloads
-
- 45
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 45
- citations for umbrella DOI https://doi.org/10.7554/eLife.38874






