Unified mechanisms for self-RNA recognition by RIG-I Singleton-Merten syndrome variants
Figures
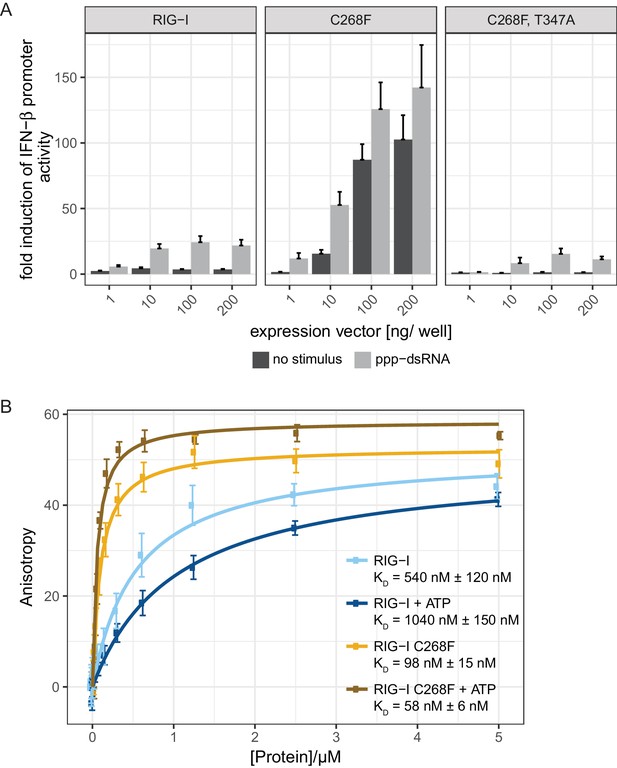
The RIG-I Singleton-Merten syndrome variant C268F signals in response to endogenous dsRNA.
(A) Fold change of interferon (IFN)-β promoter-driven luciferase activity in uninfected HEK293T RIG-I KO cells or in cells stimulated with a 19mer 5’-triphosphate (ppp)-dsRNA upon overexpression of different RIG-I mutants. Cells were co-transfected with RIG-I expression vectors and p-125luc/pGL4.74 reporter plasmids, and stimulated with ppp-dsRNA 6 hr post transfection. Firefly luciferase activities were determined in respect to Renilla luciferase activities 16 hr after RNA stimulation. All ratios were normalized to an empty vector control. n = 4–12, error bars represent mean values + standard error of the mean (SEM). (B) Fluorescence anisotropy changes measured after titrating RIG-I or RIG-I C268F in the presence or absence of ATP into solutions containing a fluorescently labeled 14mer dsRNA. All binding curves were fit to a one-site binding equation using R. n = 4, error bars represent mean values ± standard deviation (SD).
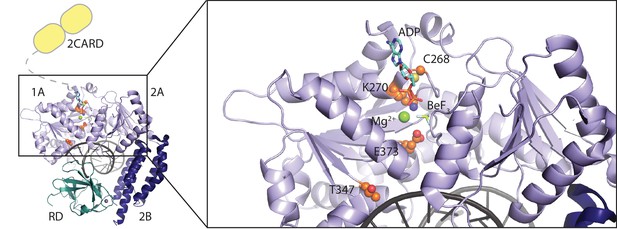
Location of RIG-I amino acid substitutions used in Figure 1.
A RIG-I-RNA-ADP·BeF3 structure (PDB: 5E3H) served as scaffold (Jiang et al., 2011). The RIG-I SF2 sub-domains (1A, 2A and 2B) are colored in light blue and dark blue. The RD is depicted in cyan and 2CARD is indicated in yellow. Mutated amino acid side chains are depicted in orange. C268 and K270 are located in the SF2 motif I (‘P-loop’) and mutation of K270 reduces ATP binding (Rawling et al., 2015). Mutation of motif II E373 slows down ATP hydrolysis but keeps the molecule's ATP-binding properties intact. T347 recognizes the RNA backbone and its mutation impedes RNA-dependent signaling of wild type RIG-I (Lässig et al., 2015). C268 and E373 are mutated in atypical Singleton-Merten syndrome.
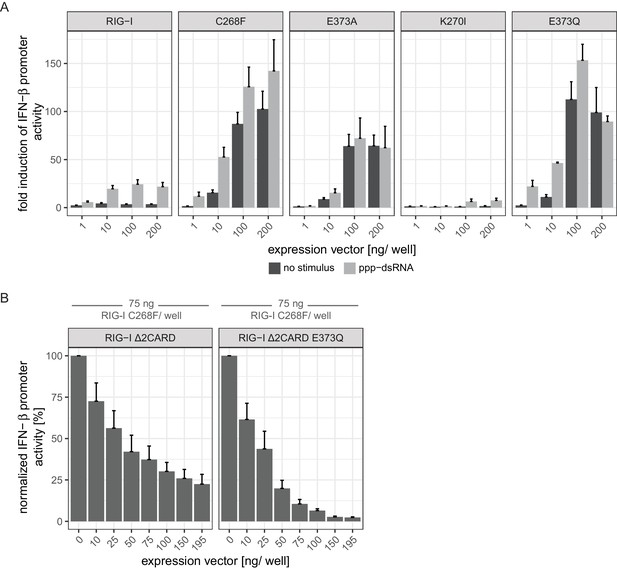
Comparison of the autoimmune signaling activity of RIG-I Singleton-Merten syndrome variants.
(A) Fold change of interferon (IFN)-β promoter-driven luciferase activity in uninfected HEK293T RIG-I KO cells or in cells stimulated with a 19mer 5’-triphosphate (ppp)-dsRNA upon overexpression of different RIG-I mutants. Cells were co-transfected with RIG-I expression vectors and p-125luc/pGL4.74 reporter plasmids, and stimulated with ppp-dsRNA 6 hr post transfection. Firefly luciferase activities were determined in respect to Renilla luciferase activities 16 hr after RNA stimulation. All ratios were normalized to an empty vector control. n = 4–12, error bars represent mean values + SEM. Overall, our results corroborate the findings reported by Jang et al. (2015) and by Lässig et al. (2015) in a concentration-dependent manner. (B) Normalized IFN-β promoter-driven luciferase activity in uninfected HEK293T RIG-I KO cells that were co-transfected with the RIG-I SMS variant C268F, p-125luc/pGL4.74 reporter plasmids and increasing amounts of signaling-incompetent versions of RIG-I. Firefly luciferase activities were determined in respect to Renilla luciferase activities 16 hr after second transfection. All ratios were normalized to the RIG-I C268F-only control. n = 4, error bars represent mean values + SEM.
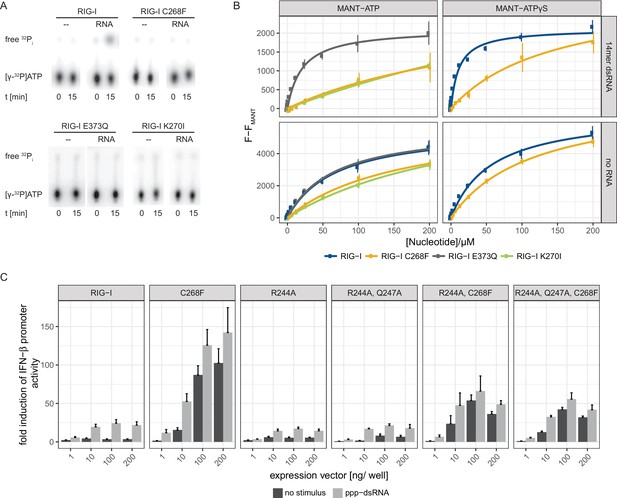
The RIG-I Singleton-Merten syndrome variant C268F is catalytically dead and has reduced ATP-binding-properties.
(A) ATP hydrolysis activity of RIG-I, the RIG-I Singleton-Merten syndrome (SMS) variant C268F and the RIG-I motif I and II mutants K270I and E373Q. RIG-I proteins were incubated with [γ-32P]-ATP in the presence or absence of a 12mer dsRNA for 15 min at room temperature and free phosphate was separated from ATP by thin layer chromatography. (B) Affinity of RIG-I, RIG-I C268F and the RIG-I motif I and II mutants to MANT-ATP or MANT-ATPγS measured by tryptophan fluorescence Förster resonance energy transfer to the MANT-nucleotide. Proteins were incubated with increasing amounts of nucleotides in the presence or absence of a 14mer dsRNA. MANT fluorescence was recorded minus a MANT-nucleotide-only control. n = 4, error bars represent mean values ± SD. (C) Fold change of interferon (IFN)-β promoter-driven luciferase activity in uninfected HEK293T RIG-I KO cells or in cells stimulated with a 19mer 5’-triphosphate (ppp)-dsRNA upon overexpression of different RIG-I mutants. Cells were co-transfected with RIG-I expression vectors and p-125luc/pGL4.74 reporter plasmids, and stimulated with ppp-dsRNA 6 hr post transfection. Firefly luciferase activities were determined in respect to Renilla luciferase activities 16 hr after RNA stimulation. All ratios were normalized to an empty vector control. n = 4–12, error bars represent mean values + SEM.
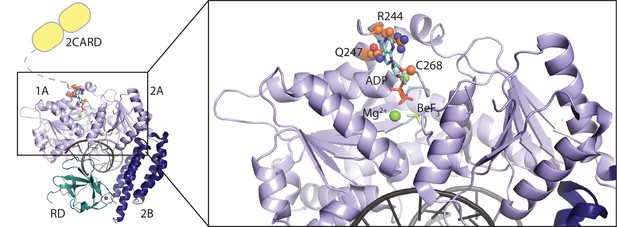
Location of RIG-I amino-acid substitutions used in Figure 2.
A RIG-I-RNA-ADP·BeF3 structure (PDB: 5E3H) served as scaffold (Jiang et al., 2011). The RIG-I SF2 sub-domains (1A, 2A and 2B) are colored in light blue and dark blue. The RD is depicted in cyan and 2CARD is indicated in yellow. Mutated amino acid side chains are depicted in orange. Q247 and R244 are located within the SF2 Q-motif and participate in ATP base recognition. C268 is mutated in atypical Singleton-Merten syndrome.
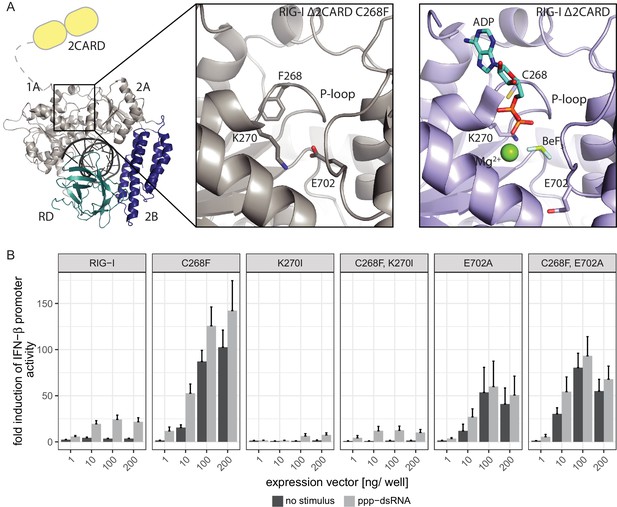
The RIG-I Singleton-Merten syndrome variant C268F induces amino acid side chain rearrangements within the active site that interfere with nucleotide binding.
(A) ATP-binding pockets of the RIG-I Singleton-Merten syndrome (SMS) variant C268F (left and middle panels) and the RIG-I wild type (right panel) bound to a 14mer dsRNA. The RIG-I SF2 sub-domains are colored in light gray or light blue (1A and 2A) and dark blue (2B). The RD is depicted in cyan and 2CARD is indicated in yellow. (B) Fold change of interferon (IFN)-β promoter-driven luciferase activity in uninfected HEK293T RIG-I KO cells or in cells stimulated with a 19mer 5’-triphosphate (ppp)-dsRNA upon overexpression of different RIG-I mutants. Cells were co-transfected with RIG-I expression vectors and p-125luc/pGL4.74 reporter plasmids, and stimulated with ppp-dsRNA 6 hr post transfection. Firefly luciferase activities were determined in respect to Renilla luciferase activities 16 hr after RNA stimulation. All ratios were normalized to an empty vector control. n = 4–12, error bars represent mean values + SEM.
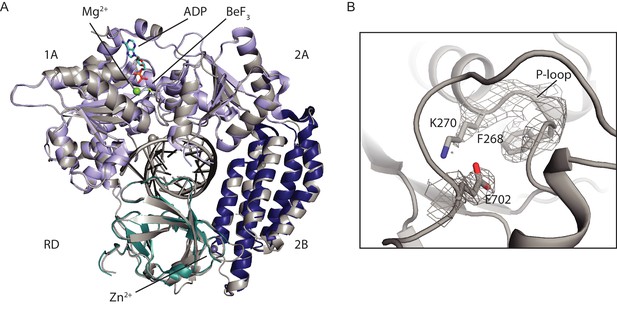
Structural comparison of the RIG-I Singleton-Merten syndrome variant C268F with wild type RIG-I.
(A) Alignment of RIG-I Δ2CARD C268F (light gray) with wild type RIG-I Δ2CARD (color code as in Figure 1—figure supplement 1) PDB 5E3H (Jiang et al., 2011). ADP·Bef3 and Mg2+ are co-crystalized with wild type RIG-I Δ2CARD but not with RIG-I Δ2CARD C268F. (B) 2Fo − Fc electron density of residues within the ATP-binding pocket of RIG-I Δ2CARD C268F at a contour level of 1σ.
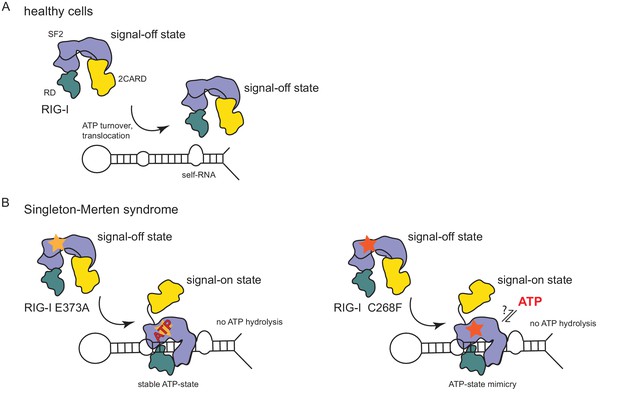
Model for the impact of Singleton-Merten syndrome mutations on self-RNA-induced RIG-I signaling.
(A) In healthy cells, wild type RIG-I occurs in a signal-off state in which 2CARD is shielded by binding to the insertion domain of SF2. Binding of RIG-I to self-RNAs is efficiently prevented through ATP-turnover-induced dissociation (for a detailed model on self- vs non-self RNA discrimination see also Lässig et al. (2015). (B) RIG-I Singleton-Merten syndrome (SMS) mutations either slow down ATP hydrolysis and stabilize the ATP-state (E373A, left side) or mimic the ATP-bound state (C268F, right side), and thus allow formation of the RIG-I signal-on state. In both cases, loss of ATP hydrolysis enhances the interaction with self-RNA and therefore results in pathogenic signaling. SMS mutations are indicated with a yellow or orange star.
Videos
Crystal structure of RIG-I Δ2CARD C268F and close-up of the active site.
The Singleton-Merten syndrome (SMS) mutation F268, as well as K270 and E702, are represented by a stick model. Theoretic locations of ADP·BeF3 and Mg2+ are indicated in faint sticks and spheres, respectively, according to a superposition with RIG-I Δ2CARD in complex with RNA and nucleotide analogue (PDB 5E3H). K270 is located at the Mg2+-binding site, whereas E702 occupies the BeF3 (ATP γ-phosphate) position.
Tables
Affinities of different RIG-I mutants to MANT-ATP or MANT-ATPγS in the presence or absence of a 14mer dsRNA. n.d., not determined, n.f., no fit possible as no saturation was reached.
https://doi.org/10.7554/eLife.38958.007| Protein | MANT-ATP | MANT-ATPγS |
|---|---|---|
| RIG-I | 72 ± 13 µM | 58 ± 7 µM |
| RIG-I + RNA | n.d. | 11 ± 1 µM |
| RIG-I E373Q | 72 ± 13 µM | n.d |
| RIG-I E373Q + RNA | 28 ± 5 µM | n.d |
| RIG-I K270I | 298 ± 81 µM | n.d |
| RIG-I K270I + RNA | n.f. | n.d |
| RIG-I C268F | 166 ± 34 µM | 116 ± 13 µM |
| RIG-I C268F + RNA | n.f. | 147 ± 55 µM |
| Reagent type (species) or source | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | HEK293T RIG-I KO | Zhu et al. (2014) | Growth in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) as monolayer | |
| Strain, strain background (Escherichia coli) | BL21 (DE3) Rosetta | Novagene | ||
| Strain, strain background (Escherichia coli) | DH10multiBac | GenevaBiotech | ||
| Strain, strain background (Spodoptera frugipeda) | Sf21 insect cells | Thermo Fisher Scientific | 11497013 | Growth in SF-900 III serum-free medium |
| Strain, strain background (Trichoplusia ni) | High Five insect cells | Thermo Fisher Scientific | B85502 | Growth in Express Five serum-free medium supplemented with 10 mM L-glutamine |
| Recombinant DNA reagent | pcDNA5/FRT/TO | Thermo Fisher Scientific | V652020 | |
| Recombinant DNA reagent | pcDNA5/FRT/TO-FLAG/ HA-RIG-I and various mutants of the same construct | Lässig et al. (2015) and this paper | Progenitors: PCR, DDX58 (cDNA) and pcDNA5/FRT/TO | |
| Recombinant DNA reagent | p-125luc | Yoneyama et al. (1996) | Firefly luciferase controlled by an interferon-β promoter | |
| Recombinant DNA reagent | pGL4.74 | Promega | E6921 | Constitutive expression of a Renilla luciferase |
| Recombinant DNA reagent | pFBDM | Berger et al. (2004) | ||
| Recombinant DNA reagent | pFBDM-His-RIG-I and various mutants of the same construct | Lässig et al. (2015) and this paper | Progenitors: PCR, DDX58 (cDNA) and pFBDM | |
| Recombinant DNA reagent | pETM11-SUMO3GFP | EMBL Heidelberg, H. Besir | https://www.embl.de/pepcore/pepcore_services/cloning/sumo/ | |
| Recombinant DNA reagent | pETM11-SUMO3-RIG-I- Δ2CARD-C268F | This paper | Progenitors: PCR, DDX58 (cDNA) and pETM11-SUMO3GFP | |
| Sequence-based reagent | 19mer 5' triphosphate dsRNA | InvivoGen | tlrl-3prna | 1 µg/mL, 5’-pppGCAUGC GACCUCUGUUUGA-3 |
| Sequence-based reagent | 14mer dsRNA | Dharmacon | 5'-CGACGCUAGCGUCG-3' | |
| Sequence-based reagent | Cy3-hpRNA | Biomers | 5'-Cy3-CCACCCGCCCCCCUAGU GAGGGGGGCGGGCC-3' | |
| Chemical compound, drug | Lipofectamine 2000 | Thermo Fisher Scientific | 11668019 | Used at 2.5x excess compared to RNA/DNA mass |
| Chemical compound, drug | MANT-ATP | Jena Bioscience | NU-202 | |
| Chemical compound, drug | MANT-ATPγS | Jena Bioscience | NU-232 | |
| Chemical compound, drug | [γ-32P]ATP | Hartmann Analytic | SRP-301 | 10 nM spiked with 3 mM unlabeled ATP |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | E1910 | |
| Software, algorithm | XDS, XSCALE | Kabsch (2010) | http://xds.mpimf-heidelberg.mpg.de/ | |
| Software, algorithm | PHASER | McCoy et al. (2007); Winn et al. (2011) | http://www.ccp4.ac.uk/ | |
| Software, algorithm | Coot | Emsley et al. (2010) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ | |
| Software, algorithm | PHENIX | Afonine et al. (2012) | https://www.phenix-online.org/ | |
| Software, algorithm | Pymol | Schrödinger | https://pymol.org/2/ | |
| Software, algorithm | R | R Development Core Team (2013) | https://www.r-project.org/ |
Data collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Crystal 1 | Crystal 2 | |
|---|---|---|
| PDB code | 6GPG | |
| Data collection | ||
| Space group | P212121 | P6522 |
| Wavelength (Å) | 1.00 | 1.00 |
| Cell dimensions | ||
| a, b, c (Å) | 112.1, 177.1, 314.8 | 175.6, 175.6, 109.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution range (Å) | 47.2–3.3 (3.42–3.30) | 46.4–2.9 (3.00–2.89) |
| Rmerge (%) | 14.3 (112) | 7.6 (206) |
| I/σI | 8.45 (1.28) | 19.72 (1.15) |
| CC1/2 | 99.8 (67.6) | 99.9 (99.7) |
| Completeness (%) | 95.3 (79.7) | 99.7 (97.4) |
| Redundancy | 3.38 (2.91) | 13.09 (13.21) |
| Refinement | ||
| Resolution (Å) | 3.3 | 2.9 |
| No. reflections | 90,121 | 22,649 |
| Rwork/ Rfree | 22.8/28.3 | 21.4/25.9 |
| No. atoms | ||
| Macromolecules | 35,730 | 5,810 |
| Ions | 10 | 2 |
| Ramachandran statistics | ||
| Favoured (%) | 92.78 | 92.71 |
| Allowed (%) | 6.39 | 6.98 |
| Outliers (%) | 0.83 | 0.31 |
| R.M.S deviations | ||
| Bond lengths (Å) | 0.011 | 0.009 |
| Angles (°) | 1.48 | 1.43 |
| B-factors | ||
| Macromolecules | 109.98 | 139.89 |
| Ions | 105.23 | 121.74 |
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.38958.013





