miR-34a is a microRNA safeguard for Citrobacter-induced inflammatory colon oncogenesis
Figures
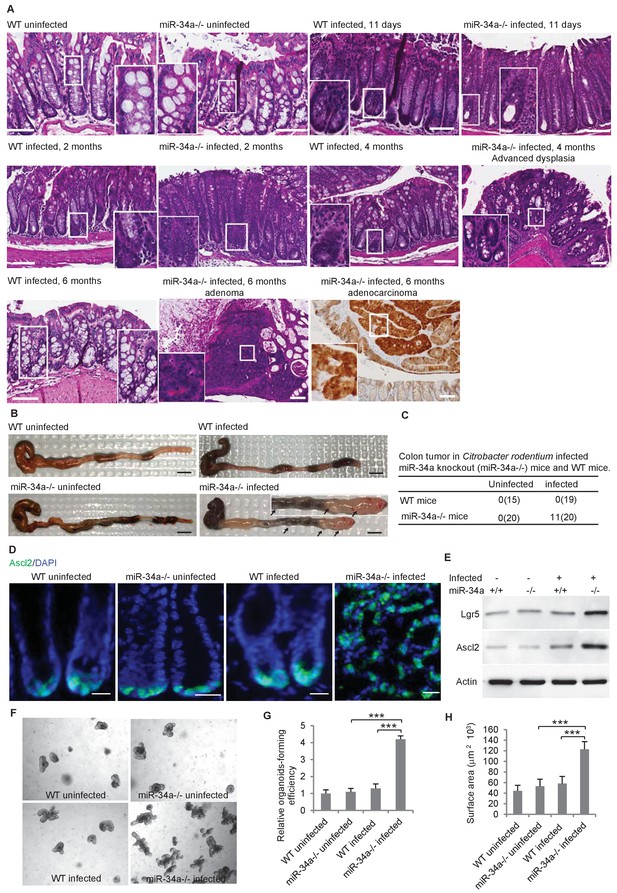
C.rodentium infection induces colonic tumor formation and stem cell enrichment in miR-34a-/- mice.
(A) Representative H and E staining of colon tissues from infected and uninfected wildtype (WT) and miR-34a-/- mice. Scale bar, 100 μm. (B) Representative images of mouse colons uninfected or infected with C. rodentium. The arrows indicate the visible colon tumors. 2 × 109 CFU C. rodentium were used to infect the mice orally. Six months after the infection, the mice were euthanized and the colons were imaged. Scale bar, 5 mm. (C) Frequencies of colonic tumor formation in infected and uninfected mice. (D) Immunofluorescence of Ascl2 showing enriched colon stem cells in miR-34a-/- colon tumors. Scale bar, 40 μm. (E) Western blot of Ascl2 and Lgr5 showing enriched colon stem cells in miR-34a-/- colon tumors. (F–H) Representative colon organoids images (F) and quantification showing organoid-forming efficiency (H) and organoid sizes. Colon organoids were cultured from uninfected mice and C. rodentium infected mice after 2 months. 1000 organoid cells from each condition were reseeded to examine organoid-forming efficiency and organoids growth.
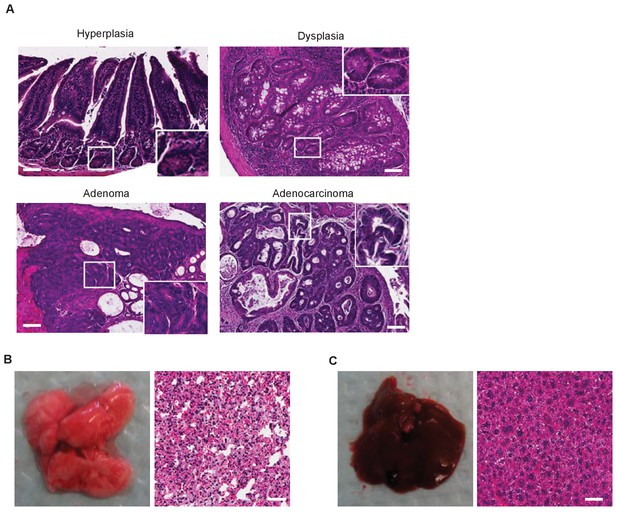
C.rodentium infection induces colonic tumor formation in miR-34a-/- mice.
(A) Representative H and E staining of C. rodentium infection-induced colon lesions in miR-34a-/- mice. (B and C) Representative images of liver (B) and lung (C) from colonic adenoma and adenocarcinoma burdened miR-34a-/- mice showing no liver or lung metastasis.
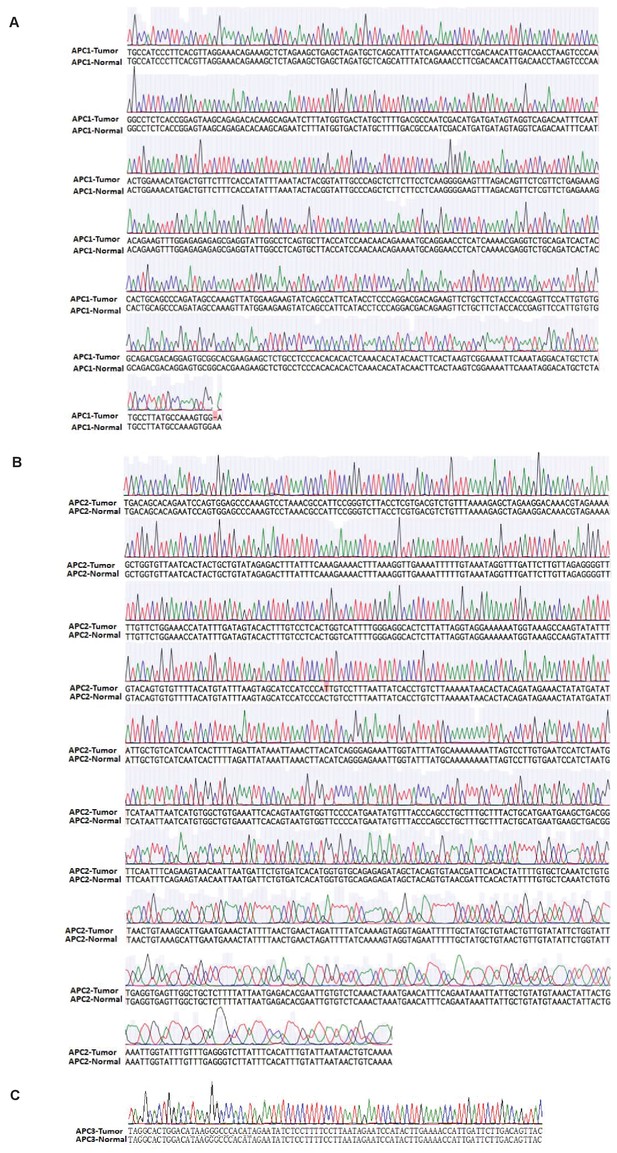
No detectable APC mutation in tumors from infected miR-34a-/- mice.
(A–C) PCR was performed using primers to amply three most APC mutation regions in mouse colon cancer. Primers: ‘GCCATCCCTTCACGTTAG’ and ‘TTCCACTTTGGCATAAGGC’ for DNA sequence containing mutation 1 (A); Primers: ‘TGACAGCACAGAATCCAGTG’ and ‘TACCAAGCATTGAGAG’ for DNA sequence contains mutation 2 (B); Primers: ‘TAGGCACTGGACATAAGGGC’ and ‘GTAACTGTCAAGAATCAATGG’ for DNA sequence containing mutation 3 (C).
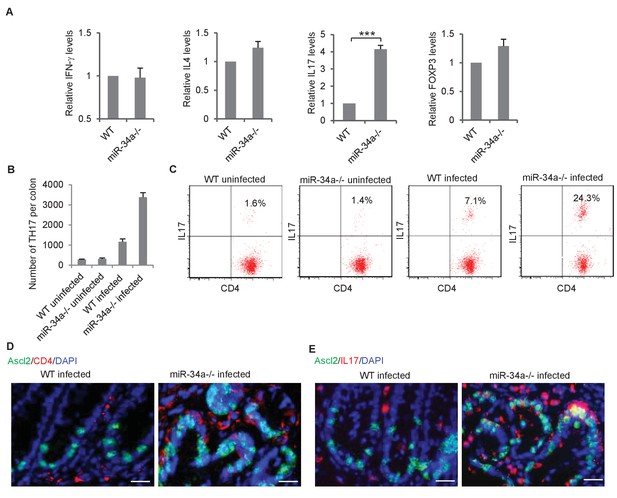
C.rodentium infection enhances Th17 cell infiltration in miR-34a-/- colonic tumors.
(A) RT-qPCR showing relative expression of the CD4+ T lymphocyte genes associated with Th1 (IFN-γ), Th2 (IL-4), Th17 (IL-17), and Treg (FOXP3) cells in the colons from C. rodentium infected wild-type and miR-34a-/- mice. (B) FACS analysis showing Th17 cells (CD4+/IL-17+) numbers in infected and uninfected mice colon at month 2. (C) FACS analyses of CD4 +T cells from each infected and uninfected WT and miR-34a-/- mice colon at month 6. The percentages of Th17 cells (CD4+/IL-17+) are marked. (D and E) Immunofluorescence of CD4 (D) and IL-17 (E) in infected colons showing enhanced Th17 cells infiltrating in miR-34a-/- colonic tumors. Scale bar, 40 μm. Error bars denote s.d. of triplicates. ***p<0.001. p-value was calculated based on Student’s t-test.
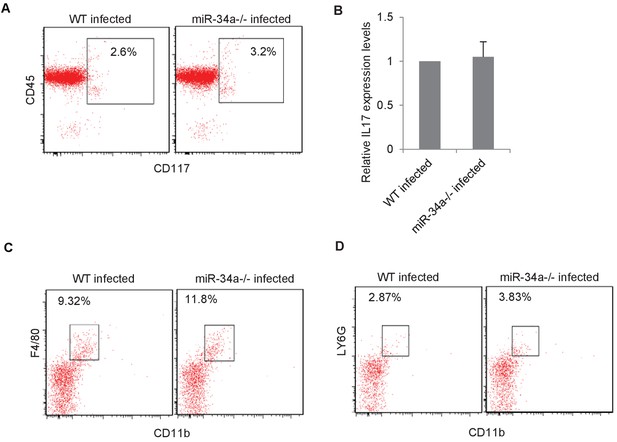
Presence of innate immune cells in C. rodentium-infected wild-type and miR-34a-/- mice.
(A) FACS plots based on CD117 and CD45 markers performed on lineage- cell populations isolated from colons of C. rodentium-infected wild-type and miR-34a-/- mice. (B) RT-qPCR showing IL-17 expression in lineage-/CD117+/CD45 +cells isolated from C. rodentium-infected colon of wild-type and miR-34a-/- mice. (C) FACs analysis of macrophages in C. rodentium infected colons of wildtype and miR-34a-/- mice. (D) FACs analysis of neutrophils in C. rodentium infected colons of wildtype and miR-34a-/- mice.
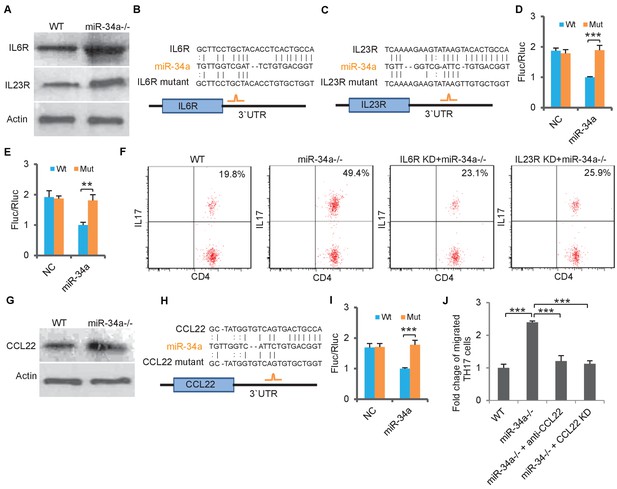
miR-34a targets IL-6R, IL-23R and CCL22.
(A) Western blot showing IL-6R and IL-23R expression levels in CD4 +T cells isolated from C. rodentium infected colon of wild-type and miR-34a-/- mice. (B and C) Schematic representation of mouse IL-6r (B) and IL-23r (C) 3`UTRs containing the putative miR-34a binding sites. (D and E) Luciferase reporter assays confirming the miR-34a binding sites. 3`UTRs of mouse IL-6r (D) and IL-23r (E) containing wild-type (Wt) or mutated (Mut) putative miR-34a binding sites were cloned into the 3`UTR of firefly luciferase (Fluc). Ectopic miR-34a expression in CT26 cells downregulated luciferase in Wt cells, but not in Mut cells. Fluc signals were normalized by a simultaneously delivered Renillar luciferase (Rluc) expression plasmid. (F) FACS showing knockdown of IL-6r or IL-23r in CD4 +T cells largely offsets the effect of miR-34a loss on Th17 cell differentiation. (G) Western blot showing increase of CCL22 expression in miR-34a-/- colon crypts. (H) Schematic representation of miR-34a binding site on the mouse CCL22 3`UTR. (I) Luciferase reporter assays confirming the miR-34a binding sites in mouse CCL22 3`UTR. (J) Chemotaxis assay showing knockdown of CCL22 in colon tumor organoid cells or neutralization of CCL22 with anti-CCL22 antibody suppresses Th17 cell migration to colon tumor organoid conditioned medium. Error bars denote s.d. of triplicates. **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test.
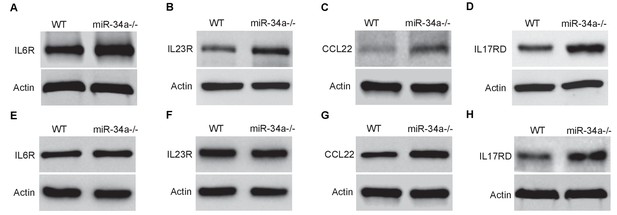
miR-34a loss upregulates target genes in uninfected mice.
(A–D) Western blots showing levels of IL-6R (A) and IL-23R (B) in CD4 +T cells and levels of CCL22 (C) and IL-17RD (D) in colon epithelial cells from wild-type (WT) and miR-34a-/- mice. (E–H) Western blot showing levels of IL-6R (E) and IL-23R (F) in CD4 +T cell and levels of CCL22 (G) and IL-17RD (H) in colon epithelial cells from Lgr5-EGFP-CreERT2/miR-34aflox/flox mice.
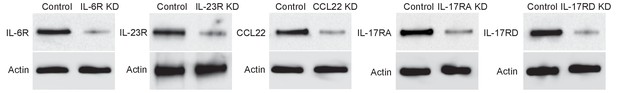
Validation of gene knockdown efficiency.
Western blots showing the knockdown efficiency of IL-6r, IL-23r, CCL22, IL-17ra, and IL-17rd.
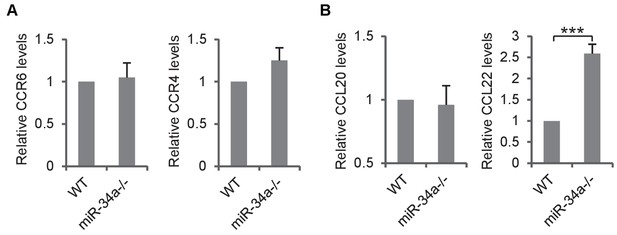
Loss of miR-34a enhances CCL22 expression in colon epithelium.
(A) RT-qPCR showing relative expression of CCR6 and CCR4 in CD4 +T cells derived from C. rodentium- infected miR-34a-/- colonic tumors and wild-type controls. (B) RT-qPCR showing relative expression of CCL20 and CCL22 in C. rodentium infected miR-34a-/- colonic tumors and wild-type controls. Error bars denote s.d. of triplicates. ***p<0.001. p-value was calculated based on Student’s t-test.
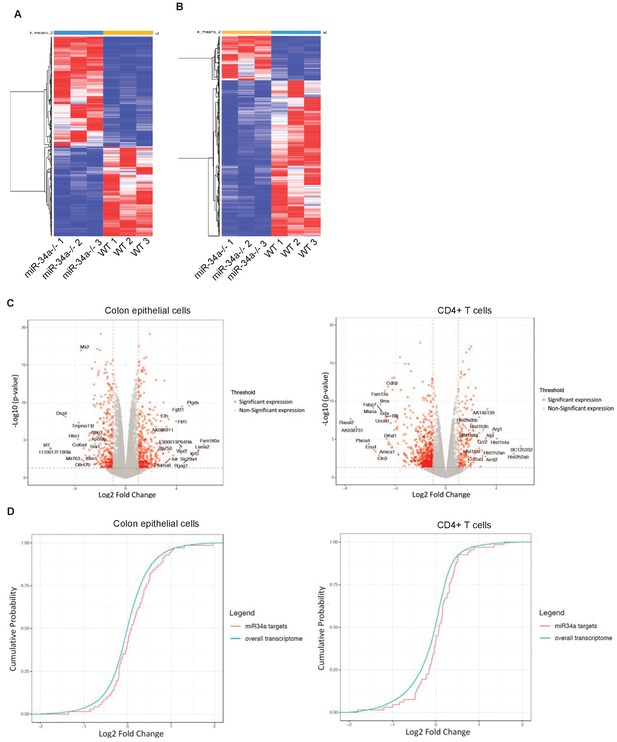
Global gene expression in colon epithelial and CD4 +T cells from wildtype vs. miR-34a-/- mice.
(A and B) Heatmaps of global gene expression in miR-34a-/- and wildtype mouse colon epithelial cells (A) and CD4 +T cells (B). (C) Volcano plots showing differential gene expression between wildtype and miR-34a-/- cells. (D) Cumulative distribution plots of miR-34a targets.
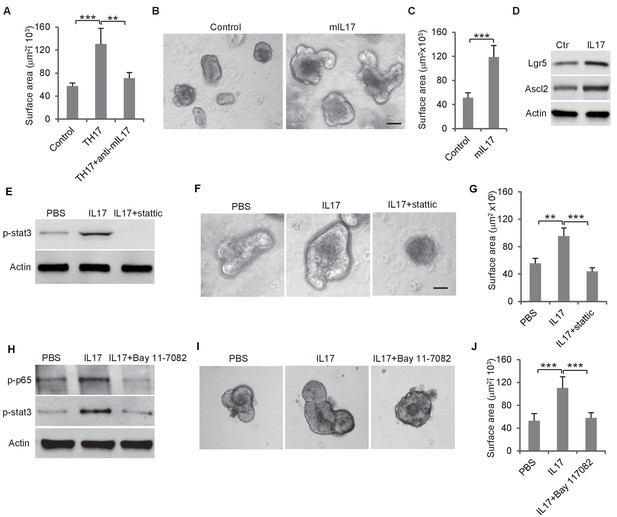
Th17 cells enhance colon organoid growth through IL-17.
(A) Th17 cells enhance colon organoid growth in co-culture. When co-cultured with Th17 cells, colon organoids grow faster with bigger surface area. Anti-IL-17 antibody abrogates Th17 promotion of colon organoids growth. (B and C) Recombinant mouse IL-17 enhances mouse organoids growth as shown by representative mouse colon organoids images (B) and quantitative organoid area (C). (D) Western blot showing that mouse IL-17 increases the expression of colon stem cell markers, Ascl2 and Lgr5, in mouse colon organoids. (E) Western blot of phospho-stat3 with IL-17 (20 ng) and STAT3 inhibitor, stattic (20 μM). (F and G) Representative organoid images (F) and quantification of organoid area (G) with IL-17 and stattic. (H) Western blot of phospho-stat3 and phospho-p65 with IL-17 (20 ng) and an NF-κB inhibitor, BAY 11–7802 (5 μM). (I and J) Representative organoid images (I) and quantificaiton of organoid area (J) with IL-17 and BAY 11–7802. Error bars denote s.d. of triplicates. **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test. .
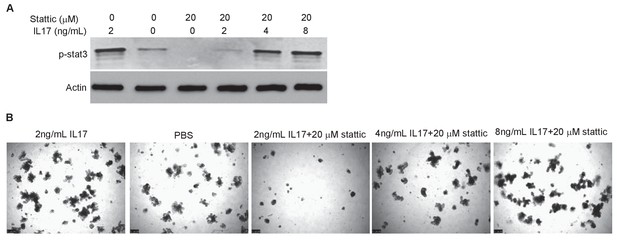
IL-17 activates STAT3 and promotes organoid growth.
(A) Western blot showing IL-17 rescues inhibition of STAT3 phosphorylation by Stattic. (B) Representative organoid images showing IL-17 rescues organoids growth suppressed by Stattic.
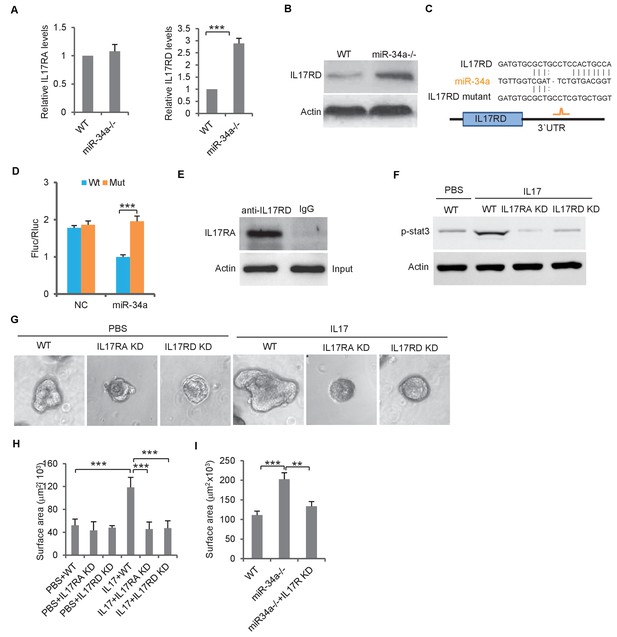
miR-34a targets orphan receptor IL-17RD in colon stem cells to suppresses IL-17-induced growth.
(A) RT-qPCR showing relative expression of IL-17ra and IL-17rd in C. rodentium infected miR-34a-/- colonic tumors and wildtype controls. (B) Western blot showing increase of IL-17RD expression in miR-34a-/- colonic tumors. (C) Schematic representation of mouse IL-17rd 3`UTR and the putative miR-34a binding site. (D) Luciferase reporter assay confirming the miR-34a binding sites in mouse IL-17rd 3`UTR. (E) Immunoprecipitation showing the IL-17RA and IL-17RD complex in the colon crypt. (F) Western blot showing IL-17RA and IL-17RD is required for IL-17 mediated STAT3 activation. (G and H) IL-17RA and IL-17RD knockdown suppresses IL-17 mediated colon organoids growth as shown by representative organoids images (G) and quantitative organoids surface area (H). (I) IL-17RD knockdown reduces miR-34a deficiency-induced colon organoids growth. Error bars denote s.d. of triplicates. **p<0.01; ***p<0.001. p-value was calculated based on Student’s t-test.
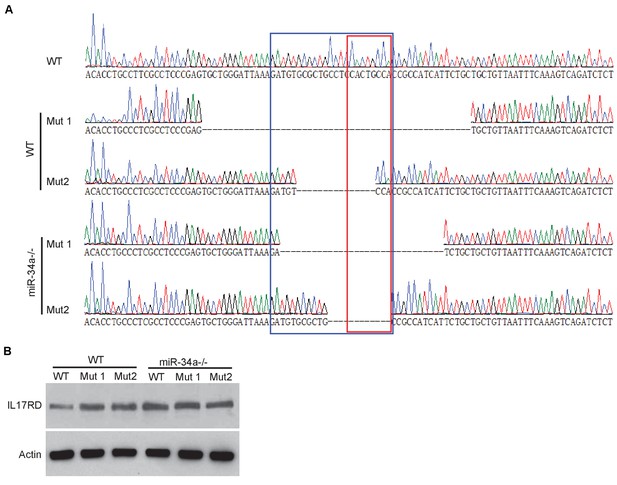
Validation of miR-34a targeting IL-17RD using CRISPER/CAS9.
(A) DNA sequencing showing mutation of miR-34a binding site in the IL-17rd 3`UTR in wildtype and miR-34a-/- colonic organoids. The blue box indicate miR-34a binding site and the red box indicates sequence complementary to the miR-34a seed region. (B) Western blot showing CRISPR-induced mutation of the miR-34a binding site increased IL-17RD expression in wildtype colon epithelial cells but not in miR-34a-/- cells.
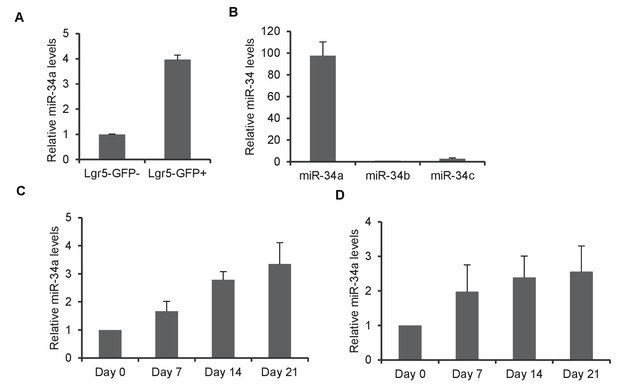
Expression of miR-34a, miR-34b and miR-34c.
(A) RT-qPCR showing higher miR-34a expression in Lgr5-GFP + colon epithelial cells than Lgr5-GFP- cells. (B) RT-qPCR showing miR-34a, miR-34b and miR-34c levels in colon epithelial cells. (C and D) RT-qPCR showing miR-34a expression levels in colon epithelial cells (C) and CD4 +T cells (D) from mice infected with C. rodentium for 7 days, 14 days and 21 days.
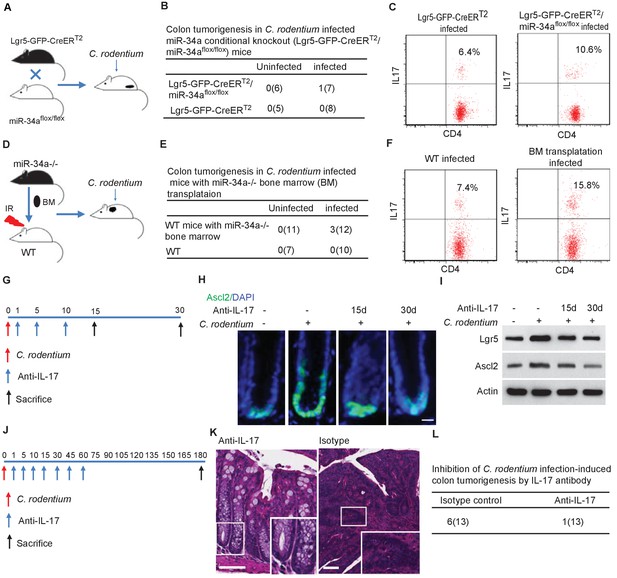
Conditional miR-34a knockout in LGR5-EGFP + stem cells and in bone marrow transplanted immune cells contribute to tumorigenesis and Th17 cell accumulation while IL-17 neutralizing antibody blocks stem cell proliferation and tumorigenesis.
(A) Schematic of the C. rodentium-infected Lgr5-EGFP-CreERT2/miR-34aflox/flox mouse model. (B) Frequencies of colon tumor formation in C. rodentium infected and uninfected Lgr5-EGFP-CreERT2/miR-34aflox/flox mice model. (C) FACS analyses of Th17 cells (CD4+/IL-17+) in C. rodentium infected and uninfected Lgr5-EGFP-CreERT2/miR-34aflox/flox mice colon. (D) Schematic of the C. rodentium-infected miR-34a-/- bone marrow transplant mouse model. (E) Frequencies of colonic tumor formation in C. rodentium infected and uninfected miR-34a-/- bone marrow transplant mice. (F) FACS analyses of Th17 cells (CD4+/IL-17+) in C. rodentium infected and uninfected miR-34a-/- bone marrow transplant mice colon. (G) Schematic of the IL-17 neutralizing antibody treatment. 500 ug isotype control or IL-17 antibody were intraperitoneally injected at the indicated days. (H) Immunofluorescence of Ascl2 showing IL-17 antibody largely abrogated C. rodentium-infection-induced colon cancer stem cell proliferation. Scale bar, 40 μm. (I) Western blot of Ascl2 and Lgr5 showing IL-17 antibody-abrogated C. rodentium-infection-induced colon stem cell marker expression. (J) Schematic of the IL-17 neutralizing antibody treatment. 500 μg isotype control or IL-17 antibody were intraperitoneally injected at the indicated days. (K) Representative H and E staining of colon tissues from IL-17 antibody or isotype control treated mice. Scale bar, 50 μm. (L) Frequencies of colonic tumor formation in IL-17 antibody or isotype control treated mice.
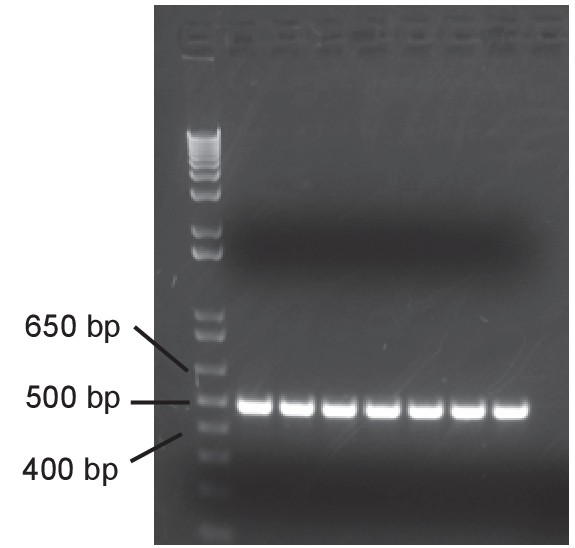
Validation of miR-34a conditional knockout mice.
Lgr5-EGFP-CreERT2/miR-34aflox/flox mice were treated with tamoxifen. Genotyping was performed to validate miR-34a deletion.
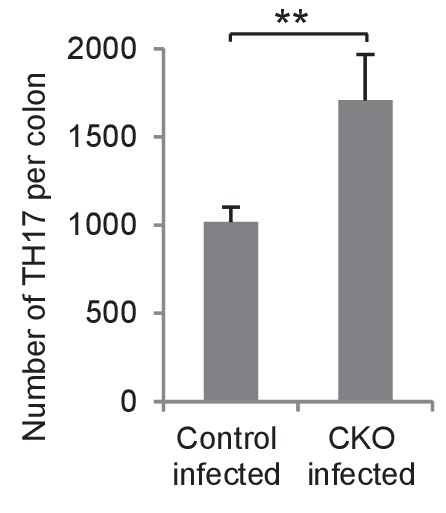
Numbers of Th17 cells in infected Lgr5-EGFP-CreERT2/miR-34aflox/flox mouse colons.
The numbers of CD4 +IL17+Th17 cells in infected Lgr5-EGFP-CreERT2 (control) and Lgr5-EGFP-CreERT2/miR-34aflox/flox mouse colon (CKO) colon, quantified by FACS analysis.
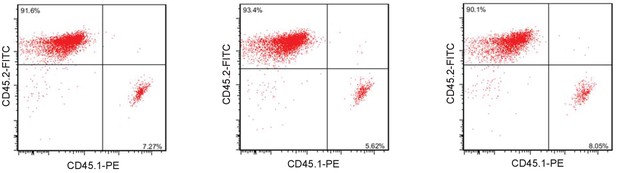
Validation of bone marrow transplantation efficiency.
Bone marrow cells from male C57BL/6J miR34a-/- donor mice with Ptprcb leukocyte marker CD45.2/Ly5.2 were transplanted into irradiated male C57BL/6J.SJL mice (Ly5.1) with the Ptprcb leukocyte marker CD45.1/Ly5.1 mice. 6 weeks after transplantation, blood cell lineages from the recipient mice were analyzed by flow cytometry using CD45.1-PE and CD45.2-FITC.
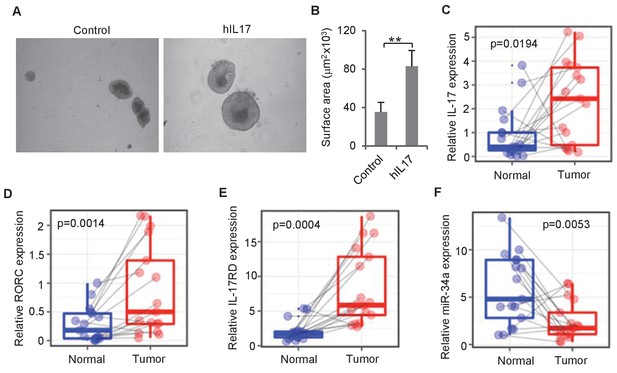
IL-17 and miR-34a expression in human CRC.
(A and B) Human IL-17 enhances human colon organoids growth as shown by representative human colon organoids images (A) and quantitative organoids area (B). Error bars denote s.d. of triplicates. **p<0.01; p-value was calculated based on Student’s t-test. (C–F) RT-qPCR of colonic tumor and normal colon tissue samples from CRC patients (Figure 7—source data 1) comparing IL-17, RORC, IL-17RD, and miR-34a transcript levels. Dots refer to different samples, and lines connect the paired samples. Error bars denote s.e.m. of 17 normal and cancer samples. p-values were calculated based on paired t-test.
-
Figure 7—source data 1
Source data for Figure 7.
This file contains the information of CRC patients.
- https://doi.org/10.7554/eLife.39479.023
-
Figure 7—source data 2
Source data for Figure 7.
This file contains RT-qPCR primers used in this study.
- https://doi.org/10.7554/eLife.39479.024
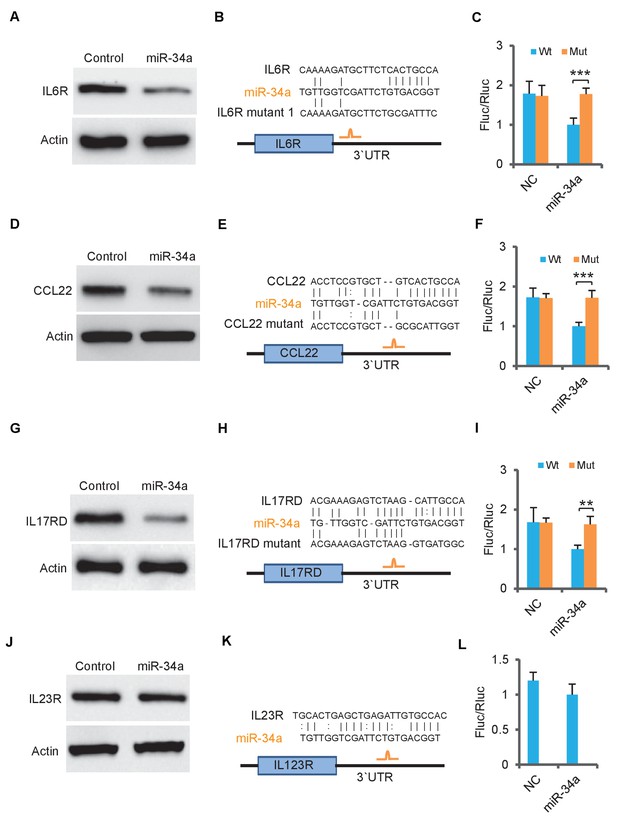
miR-34a targets human IL-6R, IL-17RD and CCL22.
(A) Western blot showing ectopic miR-34a expression suppresses human IL-6R expression. (B) Schematic representation of human IL-6r 3`UTRs containing the putative miR-34a binding sites. (C) Luciferase reporter assays confirming the miR-34a binding sites at 3`UTRs of human IL-6r. (D) Western blot showing ectopic miR-34a expression suppresses human CCL22 expression. (E) Schematic representation of human CCL22 3`UTRs containing the putative miR-34a binding sites. (F) Luciferase reporter assays confirming the miR-34a binding sites at 3`UTRs of human IL-6r. (G) Western blot showing ectopic miR-34a expression suppresses human IL-17RD expression. (H) Schematic representation of human IL-17rd 3`UTRs containing the putative miR-34a binding sites. (I) Luciferase reporter assays confirming the miR-34a binding sites at 3`UTRs of mouse IL-17rd. (J–L) Western blot and luciferase reporter assay showing human IL-23R is not targeted by miR-34a.
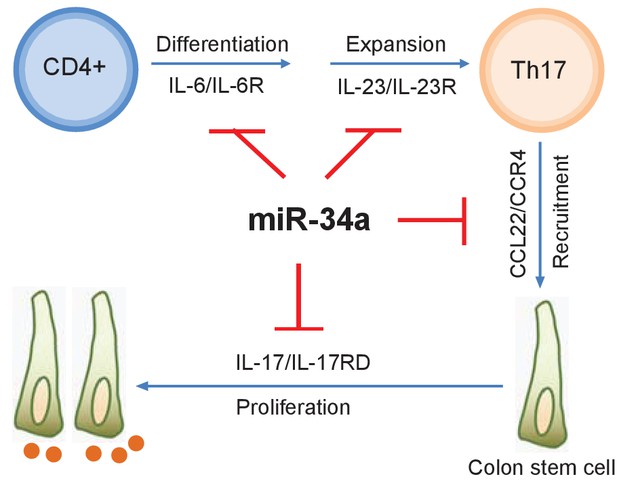
with two source data miR-34a regulates Th17 cell-mediated proliferation.
A schematic illustration of the central role of miR-34a in Th17 cell-mediated colon stem cell proliferation. miR-34a suppresses Th17 cell differentiation and expansion by targeting IL-6R and IL-23R in immune cells. miR-34a further inhibits Th17 cells recruitment by targeting CCL22 in the colon epithelium. miR-34a also inhibits IL-17RD expression to suppress IL-17-IL-17RD/IL17-RA-mediated colon stem cell proliferation.
-
Figure 8—source data 1
Transcriptomic profiling of epithelial cells.
- https://doi.org/10.7554/eLife.39479.026
-
Figure 8—source data 2
Transcriptomic profiling of CD4+ T cells.
- https://doi.org/10.7554/eLife.39479.027
Additional files
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39479.028





