CRELD1 is an evolutionarily-conserved maturational enhancer of ionotropic acetylcholine receptors
Figures
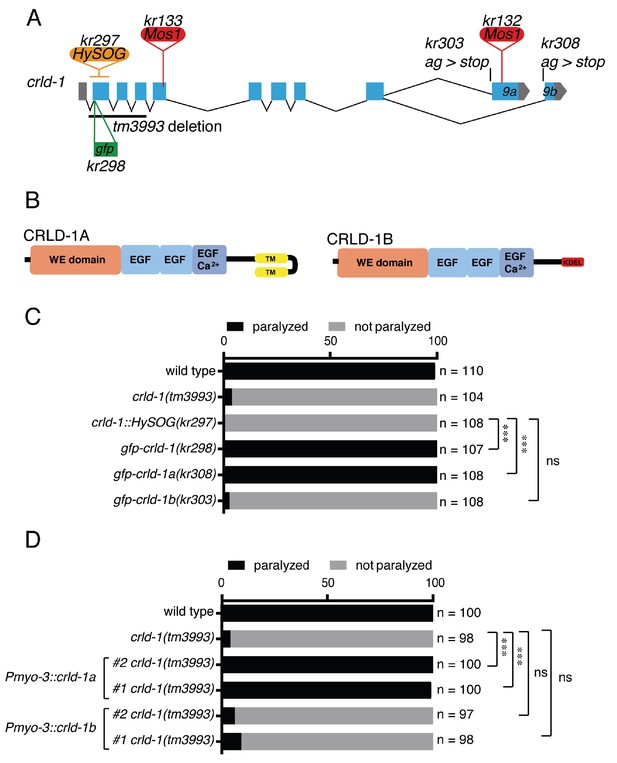
CRLD-1A isoform is sufficient for L-AChR expression based on sensitivity to levamisole.
(A) Structure of the crld-1 locus, which generates two isoforms (crld-1a and crld-1b) by alternative splicing of the last exon (exon 9a and exon 9b). The different Mos1 transposon insertions and the mutant alleles are indicated. kr303 and kr308 mutations specifically express only crld-1b and crld-1a, respectively. HySOG = hygromycinB miniSOG dual selection cassette (length = 2.8 kb). The green box indicates the position of the GFP sequence inserted in the first exon of crld-1 to generate the gfp-crld-1(kr298) knock-in allele. (B) Domain organization of CRLD-1A and CRLD-1B. SP = signal peptide, WE domain = tryptophan (W) and glutamic acid (E) enriched domain, EGF = Epidermal Growth Factor-like domain, EGF Ca2+ = Ca2+binding epidermal growth factor-like domain, TM = transmembrane domain, KDEL = Lys-Asp-Glu-Leu ER retention signal. (C) crld-1 is necessary for wild-type sensitivity to levamisole. Gray bars indicate the percentage of moving animals after overnight exposure to 1 mM levamisole, and black bars indicate the percentage of paralyzed animals. Experiments were repeated three times, n = number of animals tested. p=0,2465, ns = not significant, ***p<0.001, after Bonferroni correction, Fisher exact probability test. (D) Body wall muscle expression of crld-1a but not crld-1b rescues levamisole sensitivity in creld-1(tm3993) mutants. Gray bars indicate the percentage of moving animals after overnight exposure to 1 mM levamisole, and black bars indicate the percentage of paralyzed animals. Two independent transgenic lines were tested for each condition. Experiments were repeated four times, n = number of animals tested. p=0,2504 and 0,5369, ns = not significant, ***p<0.001, after Bonferroni correction, Fisher exact probability test.
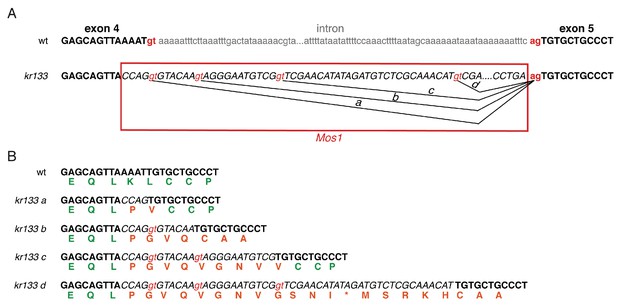
- Characterization of the kr133 mutant allele.
(A) Wild-type crld-1 and crld-1(kr133) genomic loci. Only a part of the exon 4 and exon 5 sequence is shown (bold). Regions of the intron sequence (in grey) and of the Mos1 sequence (red box) have been omitted (annotated as ‘…”). The Mos1 insertion in kr133 is located in the fourth exon of crld-1. Cryptic splice donor sites (in red) present within the Mos1 sequence are used at low frequency to generate in-frame messenger RNAs (a, b, c and d indicated by black lines). (B) Alternative crld-1 mRNAs detected by RT-PCR in crld-1(kr133) alleles (a, b, c, and d) and conceptual protein translations. The single-letter amino acid code is under the second nucleotide of the corresponding codon. Wild-type protein sequence is in green and mutated residues introduced by alternative splicing of the Mos1 transposon are in orange.
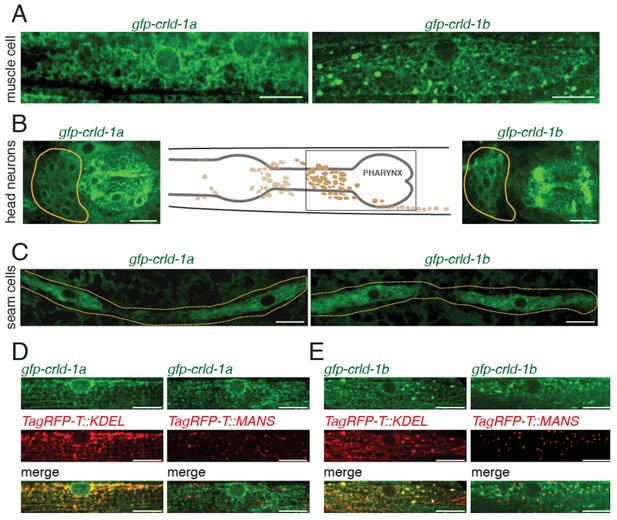
CRLD-1 is ubiquitously expressed and localizes in the ER of BWMs.
(A) Distribution of GFP-CRLD-1A (left) and GFP-CRLD-1B (right) in muscle cells of gfp-crld-1 isoform-specific knock-in worms. (B) Localization of CRLD-1A (left) and CRLD-1B (right) in the pharynx and in the lateral ganglion (encircled in yellow). The middle panel shows a schematic representation of the locations of neurons and ganglia in the head, adapted from: http://www.wormatlas.org/ver1/MoW_built0.92/nervous_system.html. (C) Localization of CRLD-1A (left) and CRLD-1B (right) in the epithelial seam cells of gfp-crld-1 isoform-specific knock-in worms. Dashed lines, seam cell outlines. (D) Expression of the ER marker TagRFP-T::KDEL in gfp-crld-1a (left) and gfp-crld-1b (right) isoform-specific knock-in strains. TagRFP-T::KDEL displays a reticular pattern throughout the cytoplasm surrounding the nucleus that co-localizes with both CRLD-1A and CRLD-1B signals. (E) CRLD-1A and CRLD-1B from gfp-crld-1a (left) and gfp-crld-1b (right) knock-in animals do not co-localize with a Golgi-resident TagRFP-T-tagged Mannosidase II protein (MANS::TagRFP-T). In (D) and (E), the Pmyo-3 promoter was used for expression of both TagRFP-T::KDEL and MANS::TagRFP-T in body wall muscles. In all panels, scale bars equal 10 μm.
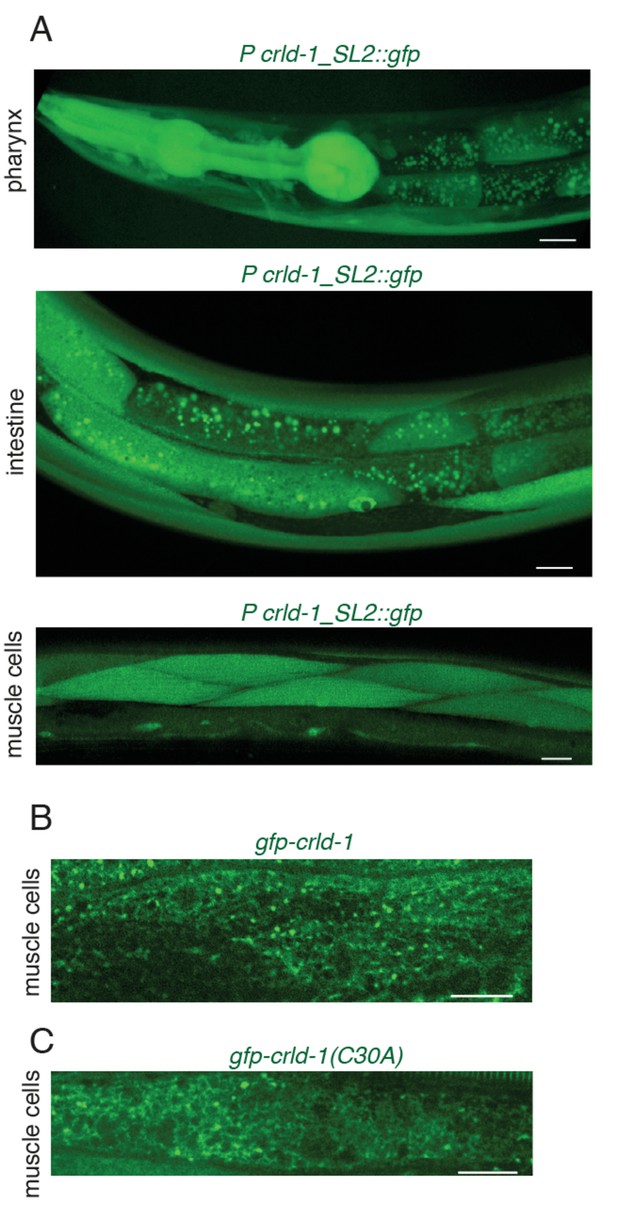
- CRLD-1 expression pattern.
(A) Confocal images of transgenic animals expressing GFP under the control of crld-1 promoter (pTB208 Pcrld-1::SL2::gfp). CRLD-1 is expressed in several tissues: pharynx, intestine and muscles. Scale bars: 10 μm. (B) Localization of CRLD-1 in muscle cells from gfp-crld-1(kr298) knock-in worms expressing both crld-1a and crld-1b isoforms. CRLD-1 reticular network is present in combination with a punctate pattern. Scale bar 10 μm. (C) Localization of CRLD-1 in muscle cells from gfp-crld-1(C30A) knock-in worms expressing both crld-1a and crld-1b isoforms. CRLD-1 reticular network is present in combination with a punctate pattern. Scale bar: 10 μm.
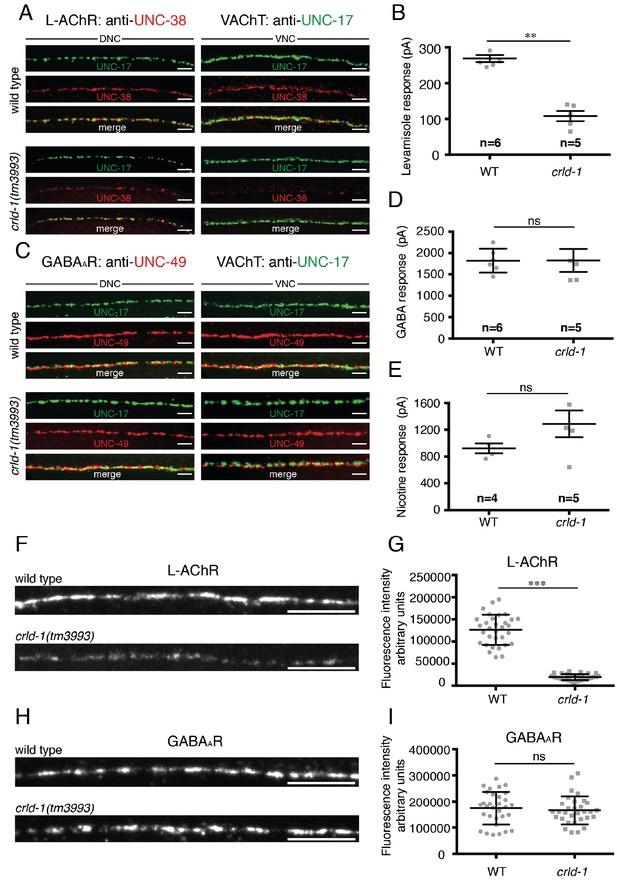
CRLD-1 is required for surface expression of L-AChRs.
(A) L-AChR expression is decreased at NMJs of crld-1(tm3993) mutants, whereas presynaptic differentiation is unaffected. L-AChRs are labeled using anti-UNC-38. Cholinergic boutons are labeled using an anti-vesicular acetylcholine transporter UNC-17 (VAChT) antibody. DNC = dorsal nerve cord, VNC = ventral nerve cord. Scale bars 10 μm. (B) Response to pressure-ejection of levamisole in voltage-clamped ventral BWMs is reduced in crld-1(tm3993). Data indicate mean ± SEM; WT: 269 ± 10 pA, n = 6 animals; crld-1(tm3993): 108 ± 14 pA, n = 5 animals; p=0.0043. Mann-Whitney test. (C) GABAAR expression is unaffected at NMJs of crld-1(tm3993) mutants compared to wild type. GABAAR are labeled using anti-UNC-49 antibodies. Cholinergic boutons are labeled using anti-UNC-17 (VAChT) antibodies. DNC = dorsal nerve cord, VNC = ventral nerve cord. Scale bars 10 μm. (D) Electrophysiological response of body-wall muscle cells to pressure-ejection of GABA in crld-1(tm3993) mutant is similar to the wild type. Data indicate mean ± SEM; WT: 1821 ± 115 pA, n = 6 animals; crld-1(tm3993): 1826 ± 270 pA, n = 5 animals; p=0.6277, ns = not significant. Mann-Whitney test. (E) Response to pressure-ejection of nicotine in body wall muscles is unaffected in crld-1(tm3993). Data indicate mean ± SEM; WT: 922 ± 76 pA, n = 4 animals; crld-1(tm3993): 1289 ± 199 pA, n = 5 animals; p=0.1905, ns = not significant. Mann-Whitney test. (F) Confocal imaging of the L-AChR reporter UNC-29::tagRFP at the ventral nerve cords of wild-type and crld-1(tm3993) mutant adult worms. Scale bars = 10 μm. (G) Quantification of UNC-29::tagRFP fluorescence at the ventral nerve cords of wild-type and crld-1(tm3993) mutant adult worms. Data indicate mean ± SD; WT: n = 32 animals; crld-1(tm3993): n = 32 animals; experiments were repeated three times.***p<0.001. Mann-Whitney test. (H) Confocal imaging of the GABAAR reporter UNC-49::tagRFP at the ventral nerve cords of wild-type and crld-1(tm3993) mutant adult worms. Scale bars = 10 μm. (I) Quantification of UNC-49::tagRFP fluorescence at the ventral nerve cords of wild-type and crld-1(tm3993) mutant adult worms. Data indicate mean ± SD; WT: n = 31 animals; crld-1(tm3993): n = 31 animals; experiments were repeated three times. p=0.4068, ns = not significant. Mann-Whitney test.
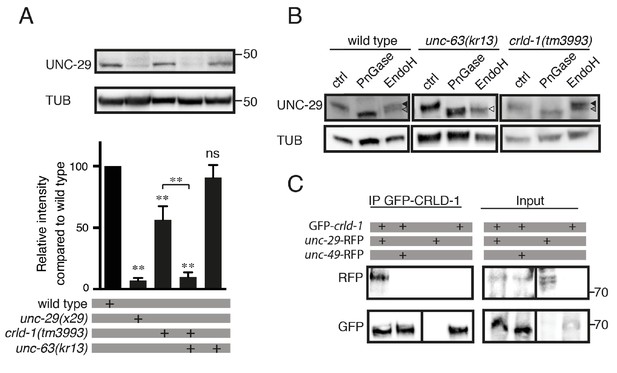
CRLD-1 is required for the stability of unassembled L-AChR subunits.
(A) L-AChR expression is reduced in crld-1(tm3993) mutants. Levels of unassembled UNC-29 L-AChR subunits detected in unc-63(kr13) are further decreased in unc-63(kr13);crld-1(tm3993) double mutants. UNC-29 levels were quantified by western blot using anti-UNC-29 antibodies and normalized to tubulin levels. Significance is indicated compared to the wild type. The significance between unc-63(kr13);crld-1(tm3993) and crld-1(tm3993) is indicated by an horizontal line. Five independent experiments were quantified (mean ± SEM). p=0.6825, ns (not significant); **p=0.0079; after Bonferroni correction, Mann-Whitney test. TUB = tubulin. (B) Remaining L-AChRs exit the ER in crld-1(tm3993). Treatments with EndoH or N-Glycosidase F (PNGase) were performed on protein extracts of mixed-stage animals before SDS-PAGE analysis. Black arrowheads indicate glycosylated forms resistant to EndoH, gray arrowheads indicate glycosylated forms partially resistant to EndoH, and white arrowheads indicate deglycosylated forms sensitive to EndoH. (C) CRLD-1 interacts with UNC-29 subunit of L-AChR in vivo. gfp-crld-1(kr298) animals were crossed with rfp-unc-29(kr208) or rfp-unc-49(kr306) to co-express CRLD-1 with RFP-tagged AChR or GABAAR subunits, respectively. Immunoprecipitation of GFP-CRLD-1 using GFP-Trap beads co-immunoprecipitated RFP-UNC-29, but not UNC-49-RFP. As a control, GFP-CRLD alone was not immunoprecipitated by anti-RFP antibody. A vertical line indicates that the lanes are not adjacent in the gel.
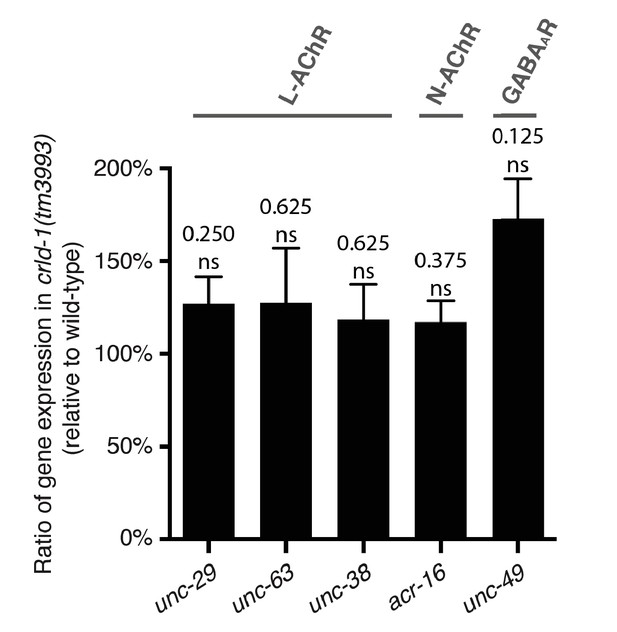
– Measurement of L-AChR subunit mRNA levels.
Transcript levels of ionotropic receptor subunits are not significantly decreased in crld-1(tm3993). Quantitative real-time PCR measurements of mRNA levels for heteromeric levamisole-sensitive AChRs (L-AChR; unc-29, unc-63, and unc-38), homomeric nicotine-sensitive AChRs (N-AChR; acr-16), and GABAA receptors (unc-49). mRNA samples were collected from synchronized L4 larvae. Fold-change, mean ± SEM in crld-1(tm3993) relative to WT is shown in four independent experiments. P values are indicated on the figure, ns (not significant), after Bonferroni Holm correction, Wilcoxon signed-rank test.
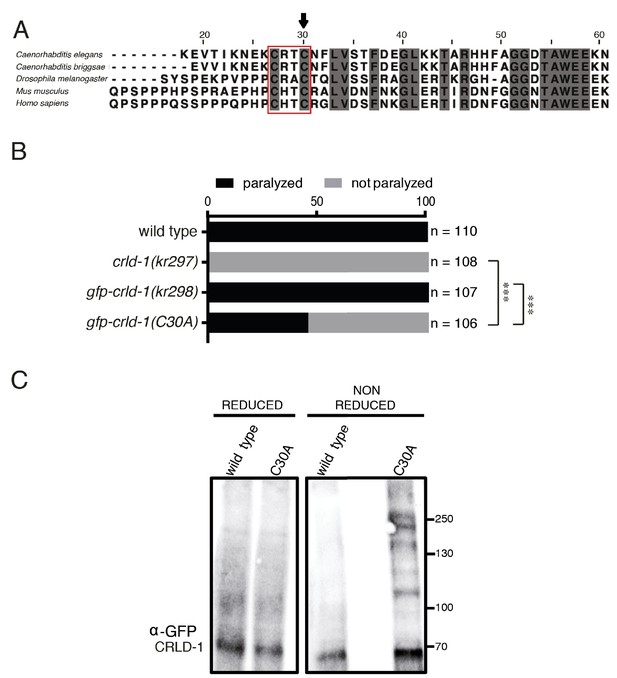
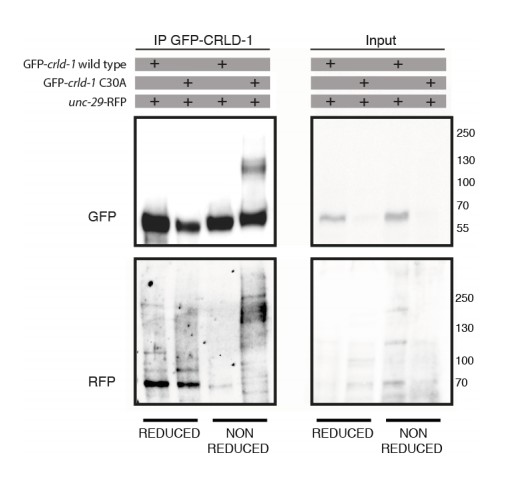
CRLD-1 displays putative PDI-like activity.
(A) ClustalO alignment of C. elegans CRLD-1 with orthologous CRLD proteins from nematodes, fly, and vertebrates. The conserved CXXC motif present in the WE domain is boxed. Identical residues conserved in all species are highlighted in dark gray. The position of the cysteine residue that was mutated to generate the C30A mutation in gfp-crld-1(C30A) knock-in worms is indicated by an arrow. (B) Animals expressing the C30A mutation in the crld-1 gene display partial levamisole resistance. Gray bars indicate the percentage of moving animals after overnight exposure to 1 mM levamisole, and black bars indicate the percentage of paralyzed animals. Experiments were repeated three times, n = number of animals tested. ***p<0.001, after Bonferroni correction, Fisher exact probability test. (C) The substrate-trapping CRLD-1 C30A mutant formed high molecular weight mixed disulphide complexes that were resolved under reducing conditions. In contrast, wild-type CRLD-1 did not form higher molecular weight complexes with putative substrate proteins. Total protein extract from gfp-crld-1(kr298) wild-type and gfp-crld-1(kr302) C30A mutant worms were separated by SDS-PAGE followed by Western blot analysis for GFP to detect CRLD-1.
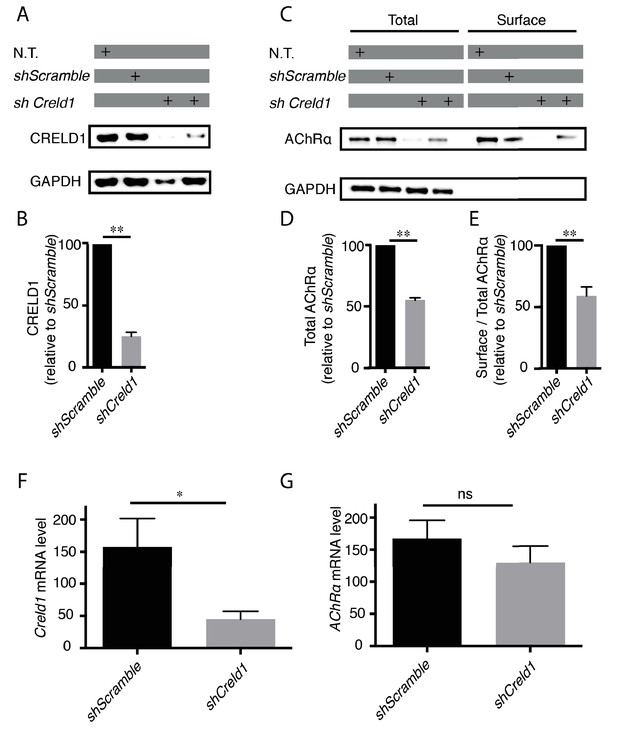
Creld1 knockdown leads to reduced AChR expression at the plasma membrane in vitro.
(A–E) Measurement of mouse CRELD1 and AChR α subunit protein levels. C2C12 cells expressing shRNA against Creld1 (shCreld1) or scrambled (shScramble) sequences were differentiated for 5 days and then subjected to surface labeling with αBT-biotin for AChRα. N.T. = non transfected cells. Streptavidin precipitates (surface) and total lysates were separated by SDS/PAGE and probed for indicated proteins (A and C). CRELD1 protein levels were reduced by 75% in cells expressing shCreld1 as compared to shScramble (B). Quantitation of total AChRα levels (D) and of the surface to total AChRα ratio (E) in shScramble (100%) and shCreld1 cells from n = 5 independent experiments. Error bars, SEM; **p=0,0079, Mann-Whitney test. (F–G) Measurement of mouse Creld1 and AChRα subunit mRNA levels. C2C12 cells expressing shScramble or shCreld1 were differentiated for 5 days and then subjected to RNA extraction. Quantitative real-time PCR measurements of mRNA levels for Creld1 (F) and AChRα subunit (G). Creld1 mRNA is decreased in shCreld1 cells compared to shScramble cells, whereas AChRα subunit mRNA is not significantly decreased in shCreld-1 cells. Mean ± SEM is shown in six independent experiments. *p=0,0411; p=0,4740, ns (not significant), Mann–Whitney test.
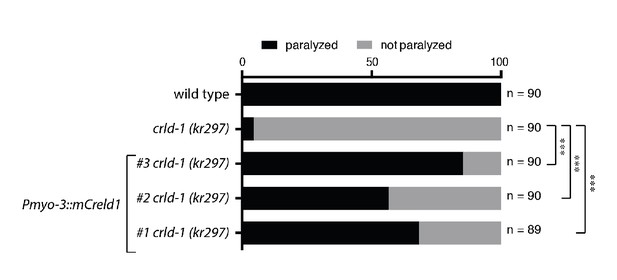
Mouse Creld1 rescues levamisole-sensitivity of crld-1 mutant worms.
Expression of mouse Creld1 in C. elegans muscle cells under the control of the Pmyo-3 promoter rescues levamisole sensitivity in kr297 mutants. Gray bars indicate the percentage of moving animals after overnight exposure to 1 mM levamisole, and black bars indicate the percentage of paralyzed animals. three independent transgenic lines were tested. Experiment was repeated three times, n = number of animals tested. ***p<0.001, after Bonferroni correction, Fisher exact probability test.
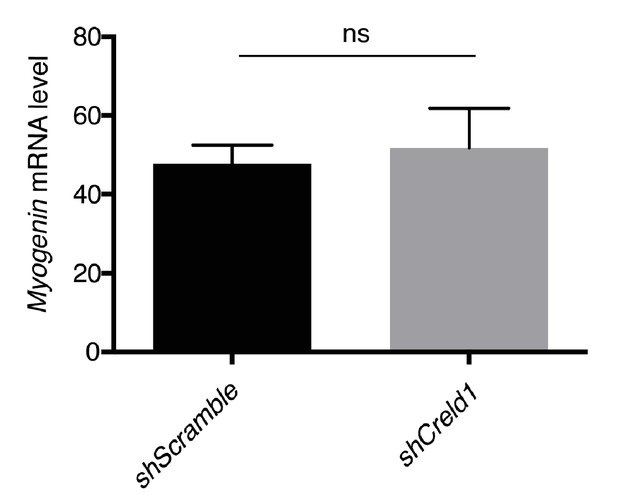
Knocking down Creld1 does not impact Myogenin transcriptional levels.
C2C12 cells expressing Scramble or Creld1 shRNA were differentiated for 5 days, and then subjected to RNA extraction. Quantitative real-time PCR measurements of mRNA levels shows that Myogenin is not significantly decreased in shCreld-1 cells. Fold-change, mean ± SEM in shCreld-1 relative to shScramble (100%) is shown in four independent experiments. p>0,9999, ns (not significant), Mann–Whitney test.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information | ||||
|---|---|---|---|---|---|---|---|---|
| Gene (C. elegans) | crld-1, F09E8.2 | this paper | WormBase ID: WBGene00 008624 | See Results, Disruption of the evolutionarily conserved gene crld-1 confers partial resistance to the cholinergic agonist levamisole. | ||||
| Gene (M. musculus) | Creld1, cysteine rich with EGF like domains 1 | PMID:12137942, PMID: 25328912, PMID: 24697899 | GeneID: 171508 | |||||
| Strain, strain background (C. elegans) | EN13 | doi:10.1534 /genetics. 104.038265 | WormBase ID:WBVar 00088264 | Strain background: N2 | ||||
| Strain, strain background (C. elegans) | ZZ29 | PMID:3668616 | WormBase ID: WBV ar00275223 | Strain background: N2 | ||||
| Strain, strain background (C. elegans) | EN208 | PMID:24896188 | WormBase ID: WBV ar02125731 | Strain background: N2 | ||||
| Strain, strain background (C. elegans) | CB407 | PMID:10377345 | WormBase ID:WBVar00143186 | Strain background: N2 | ||||
| Strain, strain background (C. elegans) | EN296: unc-49(kr296:: tagRFP) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN132: creld-1 (kr132::Mos1) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN133: crld-1 (kr133::Mos1) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2169: crld-1(tm3993) | PMID:23173093 | WormBase ID: WBVar00252554 | Strain background: N2. See M and M, Strains and Genetics | ||||
| Strain, strain background (C. elegans) | EN297: crld-1 (kr297::HySOG) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN298: crld-1 (kr298::GFP) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN302: crld-1 (kr302::GFPC30A) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN303: crld-1b (kr303::GFP) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN308: crld-1a (kr308::GFP) | this paper | Strain background: N2. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2097: crld-1 (tm3993);unc-29 (kr208::tagRFP) | this paper | Strain background: EN2169, EN308. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN4059: crld-1 (tm3993);unc-49 (kr296::tagRFP) | this paper | Strain background: EN2169, EN296. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2544: crld-1 (tm3993); krEx870 [Pmyo-3::crld-1b cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2545: crld-1(tm3993); krEx870[Pmyo-3 ::crld-1b cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2546: crld-1(tm3993); krEx870[Pmyo-3:: crld-1b cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2548: crld-1(tm3993); krEx871[Pmyo-3:: crld-1a cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2549: crld-1(tm3993); krEx871[Pmyo-3:: crld-1a cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN2550: crld-1(tm3993); krEx871[Pmyo-3:: crld-1a cDNA; myo-2::gfp] | this paper | Strain background: EN2169. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3790:crld-1(kr297); krEx1277[Pmyo-3:: mouse-creld-1 cDNA; myo-2::gfp] | this paper | Strain background: EN297. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3791:crld-1 (kr297); krEx1277 [Pmyo-3::mouse- creld-1 cDNA; myo-2::gfp] | this paper | Strain background: EN297. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3793: crld-1(kr297); krEx1277[Pmyo-3:: mouse-creld-1 cDNA; myo-2::gfp] | this paper | Strain background: EN297. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN1700: crld-1(kr132: :Mos1);krEx456 [pTB208; punc- 122::gfp] | this paper | Strain background: EN132. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3465: crld-1b(kr303:: GFP);krEx1245 [Pmyo-3::MANS:: TagRFP-T (pMR61), rol-6(su1006, panneuronal DsRed2 (pCB101)] | this paper | Strain background: EN303. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3504: crld-1b(kr303::GFP); krEx1250 [Pmyo-3:: tagRFP-T::KDEL (pMR68), rol-6(su1006, panneuronal DsRed2 (pCB101)] | this paper | Strain background: EN308. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3478: crld-1a(kr308::GFP); krEx1246 [Pmyo-3::MANS::TagRFP-T (pMR61), rol-6(su1006, panneuronal DsRed2 (pCB101)] | this paper | Strain background: EN308. See M and M, Strains and Genetics | |||||
| Strain, strain background (C. elegans) | EN3501:crld-1a(kr308:: GFP);krEx1249 [Pmyo-3::TagRFP-T::KDE;, rol-6(su1006) panneuronal DsRed2] | this paper | Strain background: EN308. See M and M, Strains and Genetics | |||||
| Cell line (M. musculus) | C2C12 mouse myoblasts | RRID:CVCL_0188 | ||||||
| Recombinant DNA reagent | MSF037586-3-CU6(OS262215) shRNA against mouse Creld1 (plasmid) | this paper | GeneCopoeia | (gccttggctactttgaggc) See M and M, Cell Culture and Western Blot | ||||
| Antibody | anti-UNC-38 | PMID: 19794415 | (1:500) | |||||
| Antibody | anti-UNC-49 | PMID: 12684444 | (1:500) | |||||
| Antibody | anti-VAChT/UNC-17 | PMID: 15457263 | (1:1000) | |||||
| Antibody | Cy3- labeled goat anti-rabbit | Jackson Immuno Research Laboratories | (1:1000) | |||||
| Antibody | A488-labeled goat anti- mouse | Molecular Probes | Cat. No.: A32723 | (1:500) | ||||
| Antibody | A488-labeled goat anti-rat | Molecular Probes | Cat. No.: A-11006 | (1:1000) | ||||
| Antibody | anti-Creld1 | Abcam | Cat. No.: ab140346 | (1:500) | ||||
| Antibody | purified mouse Anti-Acetylcholine Receptor alpha | BD Transduction Laboratories | Cat. No.: 610989 | (1:500) | ||||
| Antibody | anti GAPDH | Merck | Cat. No.: MAB374 | (1:10000) | ||||
| Antibody | Goat anti-Mouse IgG (H + L) Secondary Antibody, HRP | Thermo Fisher Scientific | Cat. No.: 62–6520 | (1:3000) | ||||
| Antibody | Goat anti- Rabbit IgG (H + L) Secondary Antibody, HRP | Thermo Fisher Scientific | Cat. No.: 65–6120 | (1:3000) | ||||
| Antibody | anti-UNC-29 | PMID:23431131 | (1:1000) | |||||
| Antibody | anti-TUBULIN | Sigma | Cat. No.:T9026- 2ML | (1:1000) | ||||
| Antibody | mouse anti-GFP | Roche | Cat. No.: 11814460001 | (1:1000) | ||||
| Antibody | RFP mouse monoclonal | ThermoFisher Scientific | Cat. No.:MA5- 15257 | (1:1000) | ||||
| Recombinant DNAreagent | pTB205:Pmyo-3::crld-1a cDNA | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pTB206: Pmyo-3:: crld-1b cDNA | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pTB208: 4,6 kb genomic fragment containing crld-1 and upstream regulatory regions fused to SL2-GFP | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMR61: Pmyo-3::RFP::MANS | PMID:23431131 | ||||||
| Recombinant DNA reagent | pMR68: Pmyo-3::RFP::KDEL | PMID:23431131 | ||||||
| Recombinant DNA reagent | pMD20: Pmyo-3:: mouse-creld-1 cDNA. | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD1: Pcrld-1:: HySOG-crld-1:: unc-54 3’UTR. | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pHZ34: Pcrld-1 ::GFP-creld-1:: unc-54 3’UTR. | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD3: this plasmid was created on the basis of pHZ34; the C30A point mutation (TGC > GCT) was introduced in the sequence of crld-1 gene. | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD5: (1 st I-SceI sgRNA) | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD7: (2nd I-SceI sgRNA) | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pPT02 | PMID: 28280212 | ||||||
| Recombinant DNA reagent | pMD8: (dpy-10 sgRNA) | PMID:25161212, this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD10: (exon9a sgRNA) | this paper | See M and M, Strains and Genetics | |||||
| Recombinant DNA reagent | pMD11: (exon9b sgRNA) | this paper | See M and M, Strains and Genetics | |||||
| Commercial assay or kit | TURBO DNA free kit, Ambion | Fisher, | Cat. No.:AM1907 | |||||
| Commercial assay or kit | iScript cDNA synthesis kit | BioRad | Cat. No.:1708891 | |||||
| Chemical compound, drug | αBT-biotin | Molecular probes/fisher | Cat. No.:B1196 | |||||
| Chemical compound, drug | Streptavidin (Sepharose Bead Conjugate) | Cell Signaling technology/ ozyme | Cat. No.:3419S | |||||
| Chemical compound, drug | levamisole | Sigma | Cat. No.:L9756 (10G) | |||||
Additional files
-
Source data 1
Source data related to Figure 1, Figure 3, Figure 4, Figure 5 and Figure 6.
- https://doi.org/10.7554/eLife.39649.013
-
Transparent reporting form
- https://doi.org/10.7554/eLife.39649.014